Overview
Welcome to LGRPv2
We've launched the Legume Genomics Research Platform version 2 (LGRPv2), now with 112 genomes including 62 legumes and Tamarindus indica. It offers browsing tools, annotations for 18,050,760 protein-coding genes, synteny analyses, and identification of paralogs, orthologs, and trait genes. New tools like DotView, SynView, DecoBrowse, and AncVisual enhance exploration. Integrated resources include a species encyclopedia, omics data, and a comparative genomics toolbox with 57 tools (29 new). Our aim with LGRPv2 is to be a comprehensive resource for legume genomics. Additionally, it offers intuitive query, analysis, and visualization tools, along with statistical charts, user manuals, communities, and submission portals, ensuring usability for all users, making it a vital asset for legume genomics research.
Portals of data
Tools quick links
|
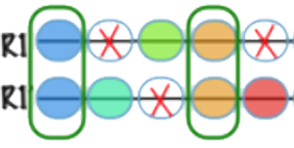
Fractionation |

|
Karyotype |
|
Gene Family |

|
Other |
|
|
|||||
|---|---|---|---|---|---|
| Species | Abbr | Chromosome number | Genes number | Data link | References |
| Abrus precatorius | Apr | 20 | 24,155 | Hovde BT et al.(2019) | |
| Aeschynomene evenia | Aev | 20 | 30,827 | Johan Ql et al.(2021) | |
| Amphicarpaea edgeworthii | Aed | 22 | 27,699 | Liu Y et al.(2020) | |
| Arachis duranensis | Adu | 20 | 33,238 | Bertioli DJ et al.(2016) | |
| Arachis hypogaea | Ahy | 40 | 65,593 | Bertioli DJ et al.(2019) | |
| Arachis ipaensis | Aip | 20 | 38,608 | Bertioli DJ et al.(2016) | |
| Arachis monticola | Amo | 40 | 69,883 | Yin D et al.(2018) | |
| Cajanus cajan | Cca | 22 | 28,222 | Parween S et al.(2015) | |
| Cicer arietinum | Car | 16 | 24,711 | Varshney et al.(2011) | |
| Dalbergia odorifera | Dod | 20 | 29,344 | Hong Z et al.(2020) | |
| Glycine max | Gma | 40 | 51,824 | Shen Y et al.(2018) | |
| Glycine soja | Gso | 40 | 46,722 | Xie M et al.(2019) | |
| Lotus japonicus | Lja | 12 | 26,152 | Li H et al.(2020) | |
| Lupinus albus | Lal | 50 | 41,277 | Hufnagel B et al.(2020) | |
| Lupinus angustifolius | Lan | 40 | 29,986 | Hane JK et al.(2016) | |
| Medicago truncatula | Mtr | 16 | 31,727 | Tang H et al.(2014) | |
| Melilotus albus | Mal | 16 | 41,853 | Wu F et al.(2021) | |
| Phaseolus vulgaris | Pvu | 22 | 27,996 | Vlasova A et al.(2016) | |
| Pisum sativum | Psa | 14 | 38,312 | Kreplak J et al.(2019) | |
| Spatholobus suberectus | Ssu | 18 | 29,597 | Qin S et al.(2019) | |
| Trifolium pratense | Tpr | 14 | 33,414 | De Vega JJ et al.(2015) | |
| Trifolium subterraneum | Tsu | 16 | 36,691 | Hirakawa H et al.(2016) | |
| Vigna radiata | Vra | 22 | 22,158 | Kang et al.(2014) | |
| Senna tora | Sto | 26 | 44,405 | Kang SH et al.(2020) | |
| Bauhinia variegata | Bva | 28 | 37,886 | Zhong Y et al.(2022) | |
| ... | ... | ... | ... | ... | ... |
Citation
If you use LGRPv2 in your scientific research, please cite us:
・Fabaceae genomes bioinformatics platform: A comprehensive database of Fabaceae genomes
・Multi-dimensional reshuffling of ancestral genome during post-polyploid diploidization shaped family Fabaceae