References & Publications
- Jeremy Schmutz#, Steven B. Cannon, Jessica Schlueter, Jianxin Ma, Therese Mitros, William Nelson, David L. Hyten, Qijian Song, Jay J. Thelen, Jianlin Cheng, Dong Xu, Uffe Hellsten, Gregory D. May, Yeisoo Yu, Tetsuya Sakurai, Taishi Umezawa, Madan K. Bhattacharyya, Devinder Sandhu, Babu Valliyodan, Erika Lindquist, Myron Peto, David Grant, Shengqiang Shu, David Goodstein, Kerrie Barry, Montona Futrell-Griggs, Brian Abernathy, Jianchang Du, Zhixi Tian, Liucun Zhu, Navdeep Gill, Trupti Joshi, Marc Libault, Anand Sethuraman, Xue-Cheng Zhang, Kazuo Shinozaki, Henry T. Nguyen, Rod A. Wing, Perry Cregan, James Specht, Jane Grimwood, Dan Rokhsar, Gary Stacey, Randy C. Shoemaker, Scott A. Jackson*. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463:178-183. 10.1038/nature08670.
- Yiming Yu#, Yidan Ouyang#, Wen Yao*. (2018). shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics 34:1229-1231. 10.1093/bioinformatics/btx763.
- Juan Yu#, Zhenhai Zhang#, Jiangang Wei#, Yi Ling#, Wenying Xu#, Zhen Su*. (2014). SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genomics 15:271. 10.1186/1471-2164-15-271.
- Eric Yao#, Robert Buels, Lincoln Stein, Taner Z Sen, Ian Holmes*. (2020). JBrowse Connect: A server API to connect JBrowse instances and users. PLoS computational biology 16:e1007261. 10.1371/journal.pcbi.1007261.
- Qihang Yang#, Tao Liu, Tong Wu, Tianyu Lei, Yuxian Li, Xiyin Wang*. (2022). GGDB: A Grameneae Genome Alignment Database of Homologous Genes Hierarchically Related to Evolutionary Events. Plant physiology 10.1093/plphys/kiac297..
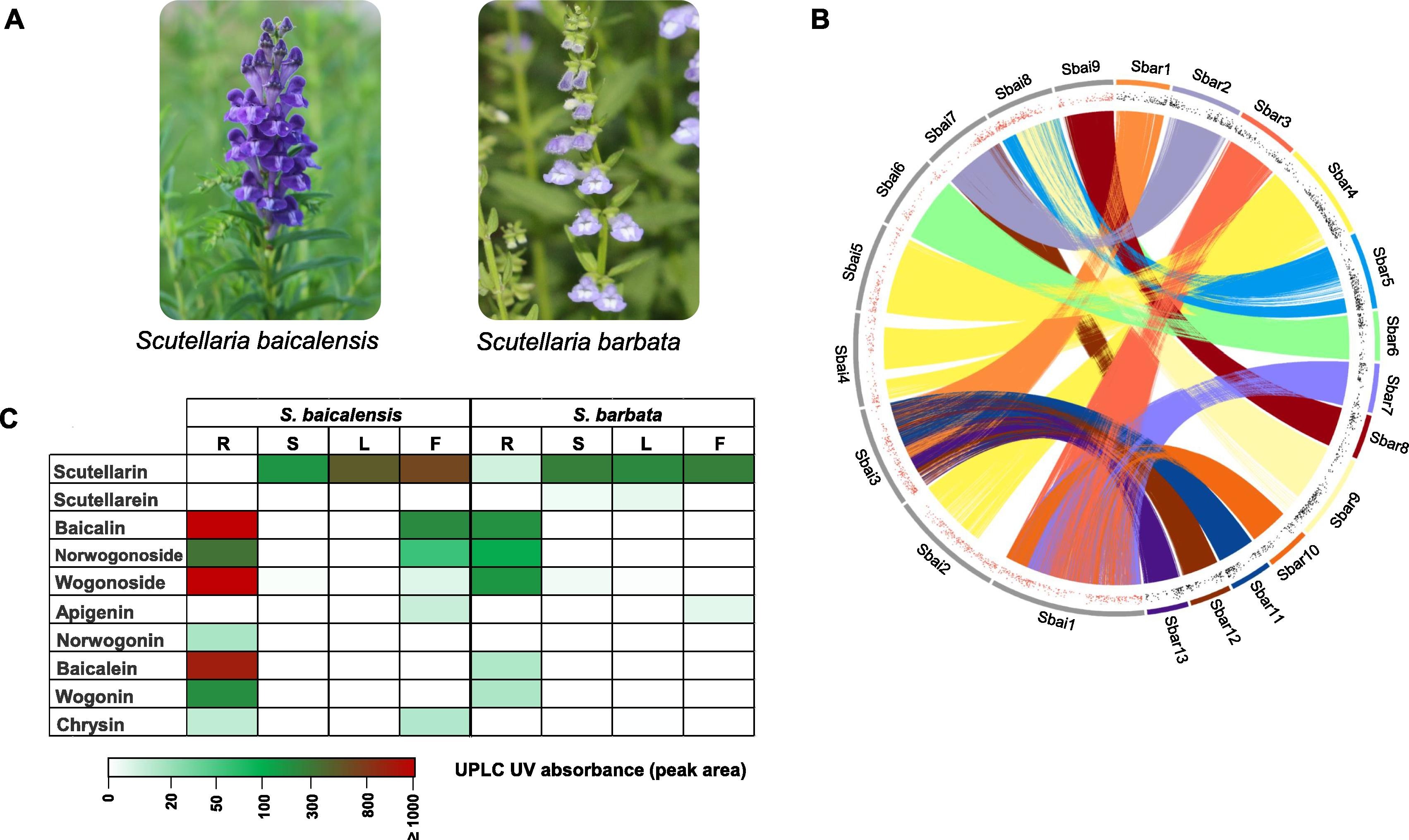
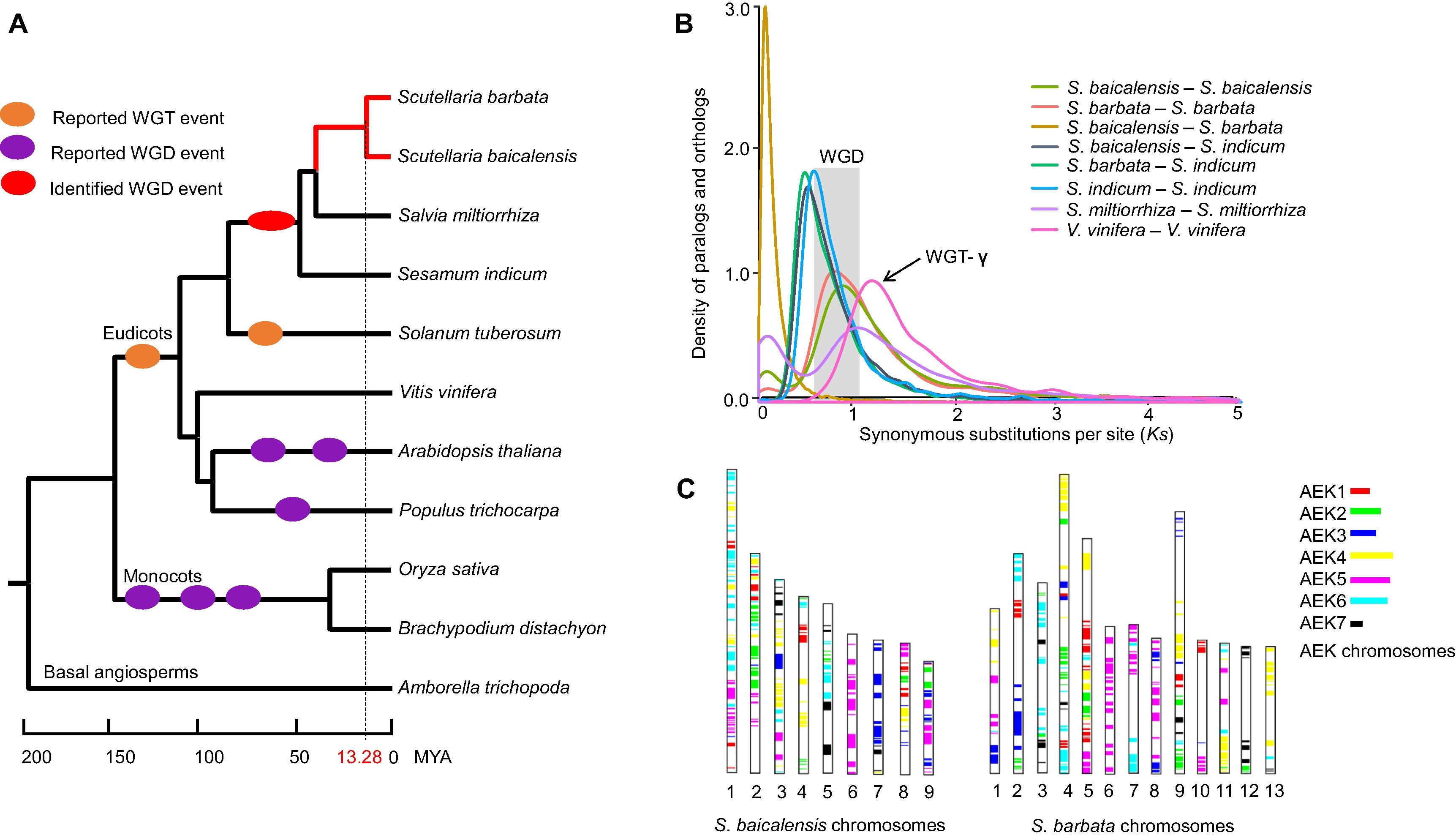
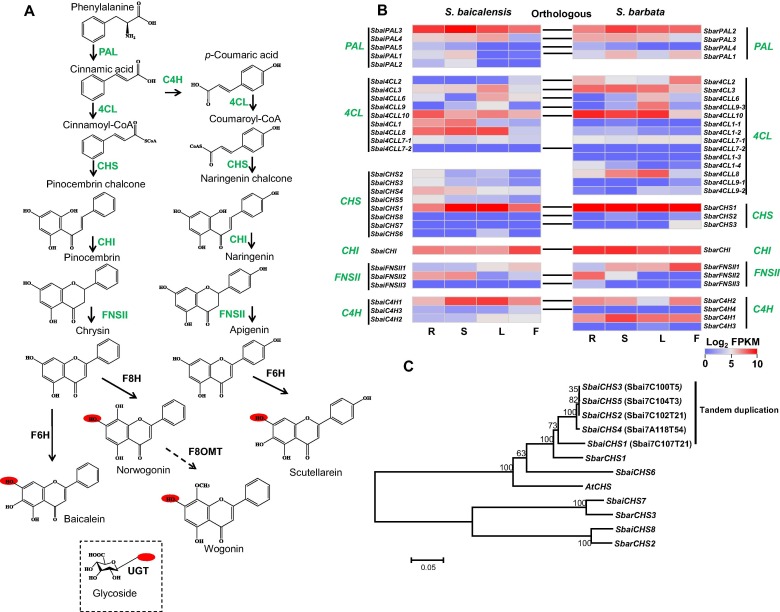
- ZhichaoXu#, RanranGao#, XiangdongPu, RongXu, JiyongWang, SihaoZheng, YanZeng, JunChen, ChunnianHe,JingyuanSong*. (2020). Comparative Genome Analysis of Scutellaria baicalensis and Scutellaria barbata Reveals the Evolution of Active Flavonoid Biosynthesis. Genomics Proteomics Bioinformatics 18:230-240. 10.1016/j.gpb.2020.06.002.
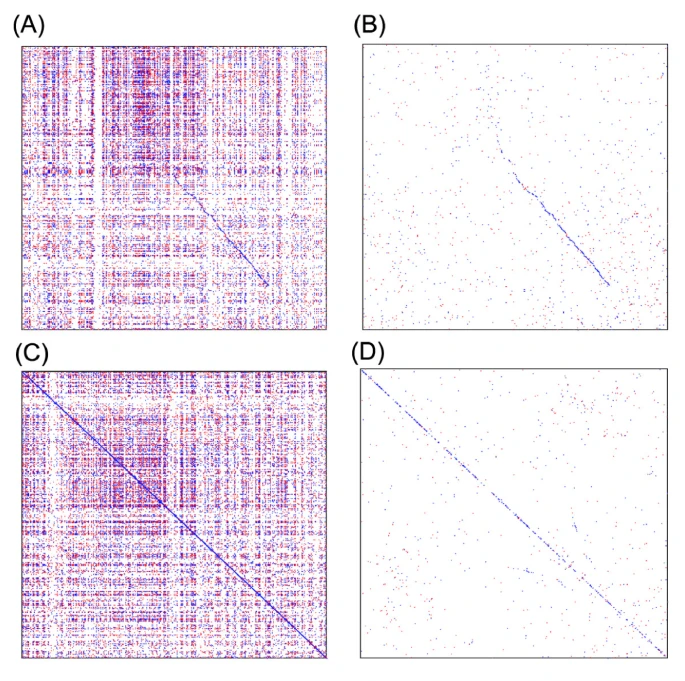
- Yupeng Wang#, Haibao Tang, Jeremy D Debarry, Xu Tan, Jingping Li, Xiyin Wang, Tae-ho Lee, Huizhe Jin, Barry Marler, Hui Guo, Jessica C Kissinger, Andrew H Paterson*. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49. 10.1093/nar/gkr1293.
- Xiyin Wang#, Xiaoli Shi, Zhe Li, Qihui Zhu, Lei Kong, Wen Tang, Song Ge, Jingchu Luo*. (2006). Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics 7:447. 10.1186/1471-2105-7-447.
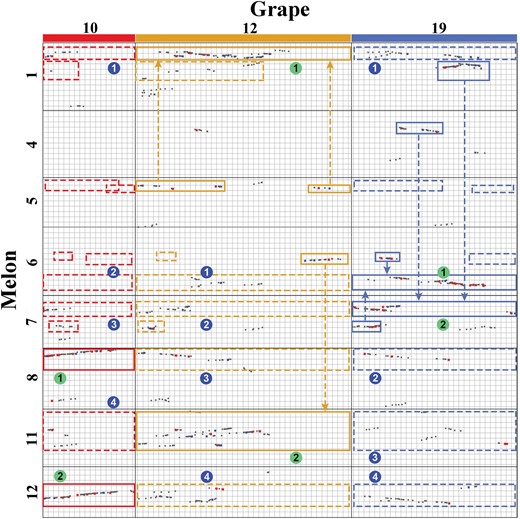
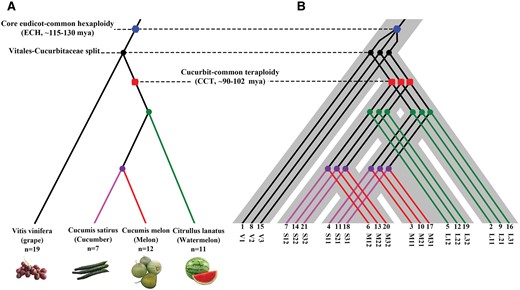
- Jinpeng Wang#, Pengchuan Sun, Yuxian Li, Yinzhe Liu, Nanshan Yang, Jigao Yu, Xuelian Ma, Sangrong Sun, Ruiyan Xia, Xiaojian Liu, Dongcen Ge, Sainan Luo, Yinmeng Liu, Youting Kong, Xiaobo Cui, Tianyu Lei, Li Wang, Zhenyi Wang, Weina Ge, Lan Zhang, Xiaoming Song, Min Yuan, Di Guo, Dianchuan Jin, Wei Chen, Yuxin Pan, Tao Liu, Guixian Yang, Yue Xiao, Jinshuai Sun, Cong Zhang, Zhibo Li, Haiqing Xu, Xueqian Duan, Shaoqi Shen, Zhonghua Zhang, Sanwen Huang, Xiyin Wang*. (2018). An Overlooked Paleotetraploidization in Cucurbitaceae. Mol Biol Evol 35:16-26. 10.1093/molbev/msx242.
- Jinpeng Wang#, Pengchuan Sun, Yuxian Li, Yinzhe Liu, Jigao Yu, Xuelian Ma, Sangrong Sun, Nanshan Yang, Ruiyan Xia, Tianyu Lei, Xiaojian Liu, Beibei Jiao, Yue Xing, Weina Ge, Li Wang, Zhenyi Wang, Xiaoming Song, Min Yuan, Di Guo, Lan Zhang, Jiaqi Zhang, Dianchuan Jin, Wei Chen, Yuxin Pan, Tao Liu, Ling Jin, Jinshuai Sun, Jiaxiang Yu, Rui Cheng, Xueqian Duan, Shaoqi Shen, Jun Qin, Meng-Chen Zhang, Andrew H Paterson, Xiyin Wang*. (2017). Hierarchically Aligning 10 Legume Genomes Establishes a Family-Level Genomics Platform. Plant physiology 174:284-300. 10.1104/pp.16.01981.
- Jinpeng Wang#, Jun Qin#, Pengchuan Sun, Xuelian Ma , Jigao Yu, Yuxian Li, Sangrong Sun, Tianyu Lei, Fanbo Meng, Chendan Wei, Xinyu Li, He Guo, Xiaojian Liu, Ruiyan Xia, Li Wang, Weina Ge, Xiaoming Song, Lan Zhang, Di Guo, Jinyu Wang, Shoutong Bao, Shan Jiang, Yishan Feng, Xueping Li, Andrew H Paterson, Xiyin Wang*. (2019). Polyploidy Index and Its Implications for the Evolution of Polyploids. Frontiers in genetics 10:807. 10.3389/fgene.2019.00807.
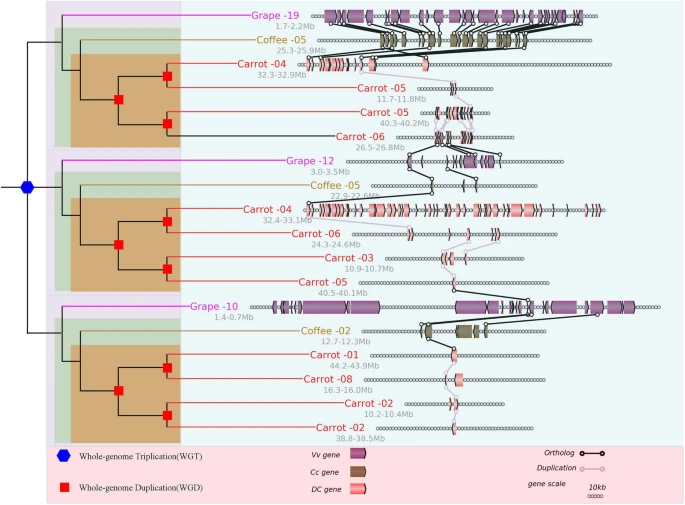
- Jinpeng Wang#, Jigao Yu, Yuxian Li, Chendan Wei, He Guo, Ying Liu, Jin Zhang, Xiuqing Li, Xiyin Wang*. (2020). Sequential Paleotetraploidization shaped the carrot genome. BMC plant biology 20:52. 10.1186/s12870-020-2235-7.
- Haibao Tang#, Vivek Krishnakumar, Shelby Bidwell, Benjamin Rosen, Agnes Chan, Shiguo Zhou, Laurent Gentzbittel, Kevin L Childs, Mark Yandell, Heidrun Gundlach, Klaus FX Mayer, David C Schwartz, Christopher D Town*. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. 10.1186/1471-2164-15-312.
- Pamela S Soltis#, D Blaine Marchant, Yves Van de Peer, Douglas E Soltis*. (2015). Polyploidy and genome evolution in plants. Current opinion in genetics & development 35:119-125. 10.1016/j.gde.2015.11.003.
- Jiaqing Yuan#, Jinpeng Wang#, Jigao Yu, Fanbo Meng, Yuhao Zhao, Jing Li, Pengchuan Sun, Sangrong Sun, Zhikang Zhang, Chao Liu, Chendan Wei, He Guo, Xinyu Li, Xueqian Duan, Shaoqi Shen, Yangqin Xie, Yue Hou, Jin Zhang, Tariq Shehzad*, Xiyin Wang*. (2019). Alignment of Rutaceae Genomes Reveals Lower Genome Fractionation Level Than Eudicot Genomes Affected by Extra Polyploidization. Frontiers in plant science> 10:986. 10.3389/fpls.2019.00986.
- Colin Ruprecht#, Rolf Lohaus#, Kevin Vanneste, Marek Mutwil, Zoran Nikoloski, Yves Van de Peer*, Staffan Persson*. (2017). Revisiting ancestral polyploidy in plants. Science advances 3:e1603195. 10.1126/sciadv.1603195.
- Renny-Byfield#, S. Wendel, J.F*. (2014). Doubling down on genomes: polyploidy and crop plants. American journal of botany 101:1711-1725. 10.3732/ajb.1400119.
- Longhui Ren#, Wei Huang#, Steven B Cannon*. (2019). Reconstruction of ancestral genome reveals chromosome evolution history for selected legume species. The New phytologist 223:2090-2103. 10.1111/nph.15770.
- Xin Qiao#, Qionghou Li, Hao Yin, Kaijie Qi, Leiting Li, Runze Wang, Shaoling Zhang*, Andrew H. Paterson*. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol 20:38. 10.1186/s13059-019-1650-2.
- Anurag Priyam#, Ben J Woodcroft, Vivek Rai, Ismail Moghul, Alekhya Munagala, Filip Ter, Hiten Chowdhary, Iwo Pieniak, Lawrence J Maynard, Mark Anthony Gibbins, HongKee Moon, Austin Davis-Richardson, Mahmut Uludag, Nathan S Watson-Haigh, Richard Challis, Hiroyuki Nakamura, Emeline Favreau, Esteban A Gómez, Tomás Pluskal, Guy Leonard, Wolfgang Rumpf, Yannick Wurm*. (2019). Sequenceserver: A Modern Graphical User Interface for Custom BLAST Databases. Mol Biol Evol 36:2922-2924. 10.1093/molbev/msz185.
- Simon C Potter#, Aurélien Luciani, Sean R Eddy, Youngmi Park, Rodrigo Lopez, Robert D Finn*. (2018). HMMER web server: 2018 update. Nucleic Acids Res 46:W200-w204. 10.1093/nar/gky448.
- Fanbo Meng#, Qiang Tang, Tianzhe Chu, Xianhai Li, Yue Lin, Xiaoming Song, Wei Chen*. (2022). TCMPG: an integrative database for traditional Chinese medicine plant genomes. Hortic Res 9:uhac060. 10.1093/hr/uhac060.
- Paola Martinez-Amador#, Nori Castañeda, Antonio Loza, Lizeth Soto, Enrique Merino, Rosa Maria Gutierrez-Rios*. (2019). Prediction of protein architectures involved in the signaling-pathway initiating sporulation in Firmicutes. BMC research notes 12:686. 10.1186/s13104-019-4712-3.
- Drishti Mandal#, Deevita Srivastava#, Senjuti Sinharoy*. (2020). Optimization of Hairy Root Transformation for the Functional Genomics in Chickpea: A Platform for Nodule Developmental Studies. Methods Mol Biol 2107:335-348. 10.1007/978-1-0716-0235-5_18.
- Fábio Madeira#, Matt Pearce, Adrian R N Tivey, Prasad Basutkar, Joon Lee, Ossama Edbali, Nandana Madhusoodanan, Anton Kolesnikov, Rodrigo Lopez*. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50:W276-279. 10.1093/nar/gkac240.