Hierarchical alignments of P. vulgaris
Inter-genomic and intra-genomic comparisons can help reveal the structural complexity of Fabaceae genomes. We used V. vinifera and P. vulgaris as references, respectively, and by comparing homologous gene locus maps and Ks values between V. vinifera and Fabaceae, P. vulgaris and other Fabaceae, we could separate orthologous and paralogous genes produced by different polyploidization in the species genomes. We created two hierarchical lists of homologous genes using V. vinifera and P. vulgaris as references, respectively. The way to establish a hierarchical list of homologous genes can be found in some previously published articles.
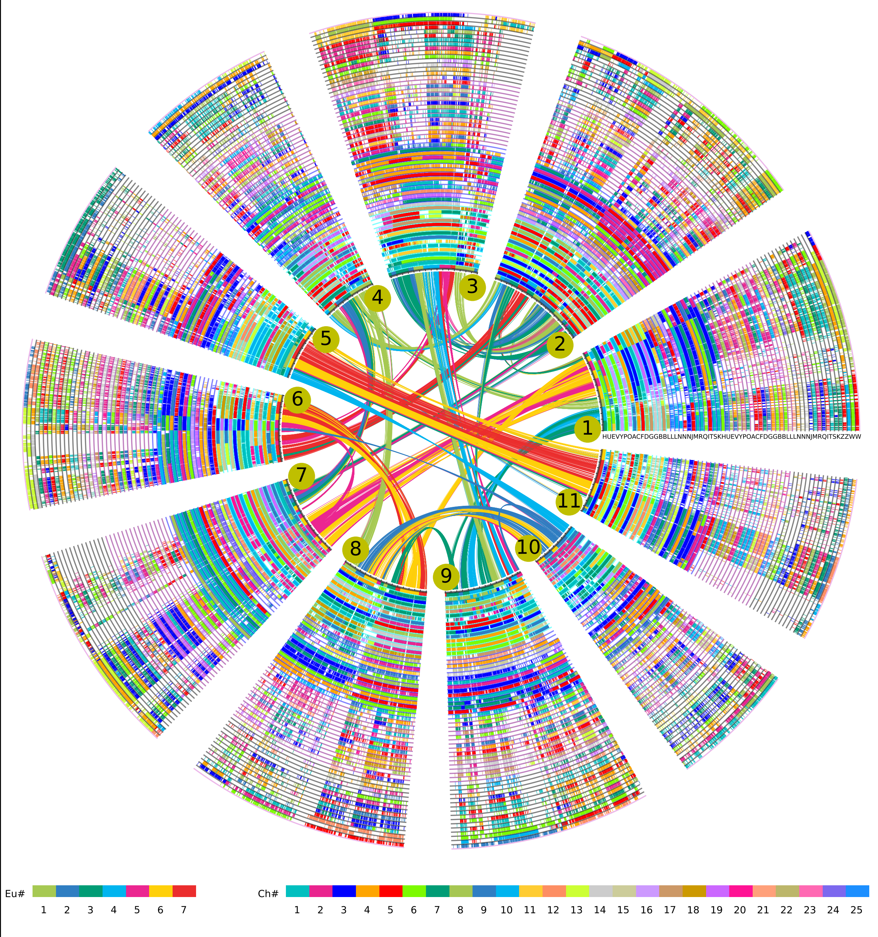
Homologous alignments of Papilionoideae, Caesalpinioideae and Cercidoideae genomes with V. vinifera as a reference. The V. vinifera chromosomes form the innermost circle, and their paralogous genes in collinearity are linked by curving lines and corresponds colored according to the 7 core-eudicot-common ancestral chromosomes inferred previously (Jaillon, et al. 2007).The V. vinifera chromosome region had 1, 2 and 2 orthologous regions in Papilionoideae, Caesalpinioideae and Cercidoideae genomes, respectively, and displayed in 65 circles, each circle represents one subgenome. Short line between two circles show colinear genes, which corresponds colors of Ch# encoded. The color scheme is shown at the bottom.
The relevant gene ids can be obtained from the Fabaceae blast and match under the Tools module.This link is Fabaceae blast and match
| Pvu1 | Adu1 | Aed1 | Aev1 | Ahy1 | Aip1 | Amo1 | Apr1 | Car1 | Cca1 | Dod1 | Gma1 | Gma1 | Gso1 | Gso1 | Lal1 | Lal1 | Lal1 | Lan1 | Lan1 | Lan1 | Lja1 | Mal1 | Mtr1 | Psa1 | Ssu1 | Tpr1 | Tsu1 | Vra1 | Pvu2 | Adu2 | Aed2 | Aev2 | Ahy2 | Aip2 | Amo2 | Apr2 | Car2 | Cca2 | Dod2 | Gma2 | Gma2 | Gso2 | Gso2 | Lal2 | Lal2 | Lal2 | Lan2 | Lan2 | Lan2 | Lja2 | Mal2 | Mtr2 | Psa2 | Ssu2 | Tpr2 | Tsu2 | Vra2 | Sto2 | Sto2 | Bva2 | Bva2 |
|---|