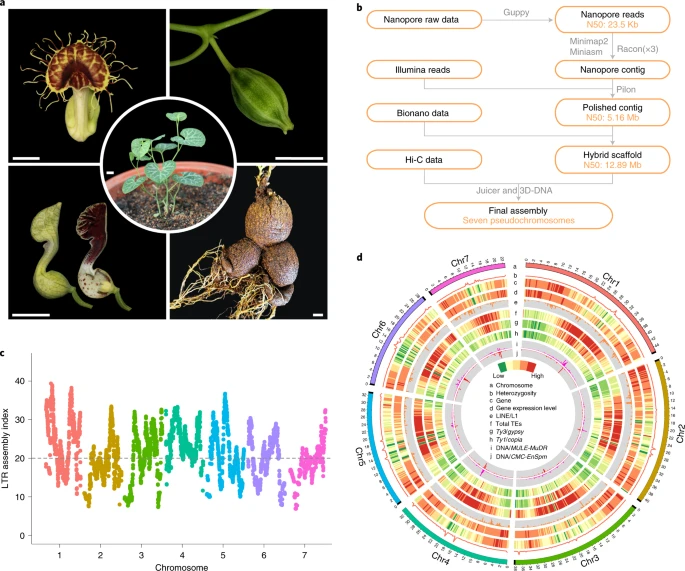
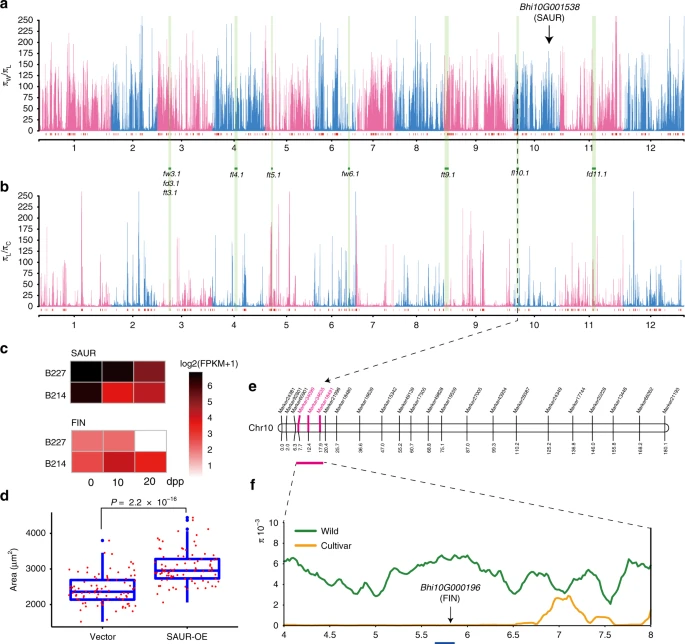
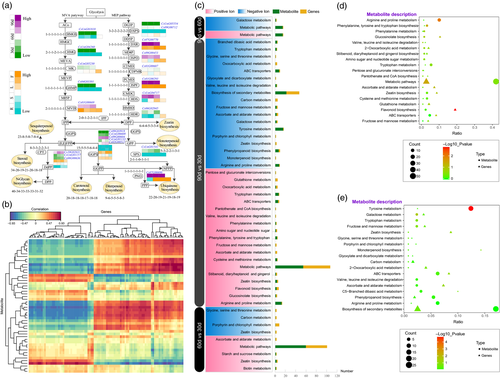
References
- Kunpeng Li#, Peng Xu#, Jinpeng Wang, Xin Yi, Yuannian Jiao*, Identification of errors in draft genome assemblies at single-nucleotide resolution for quality assessment and improvement, Nature Communications, 2023, 14:6556.
- Xueqing Yan#, Runxian Yu, Jinpeng Wang, Yuannian Jiao*, IAncestral genome reconstruction and the evolution of chromosomal rearrangements in Triticeae, J Genet Genomics, 2025, 1:S1673-8527(24)00370-9. Yan X, Yu R, Wang J, Jiao Y. Ancestral genome reconstruction and the evolution of chromosomal rearrangements in Triticeae. J Genet Genomics. 2025 Jan 1:S1673-8527(24)00370-9. doi: 10.1016/j.jgg.2024.12.017. Epub ahead of print. PMID: 39746604.
- Xiaochong Li#, Jinbin Wang#, Yanan Yu#, Jinpeng Wang, Changping Li, Zixian Zeng, Ning Li, Zhibin Zhang, Qianli Dong, Yiyang Yu, Xiaofei Wang, Tianya Wang, Corrinne E. Grover, Bin Wang, Bao Liu*, Jonathan F. Wendel*, Lei Gong* Genomic rearrangements and evolutionary changes in 3D chromatin topologies in the cotton tribe (Gossypieae), BMC Biology, 2023, 21(1):56.
- Yan Zhang#, Weina Ge#, YJia Teng#, Yanmei Yang#, Jianyu Wang#, Zijian Yu, Jiaqi Wang, Qimeng Xiao, Junxin Zhao, Shaoqi Shen, Yishan Feng, Shoutong Bao, Yu Li, Yuxian Li a, Tianyu Lei, Yuxin Pan, Lan Zhang, Jinpeng Wang*, Allotetraploidization event of coptis chinensis shared by all Ranunculales, Horticultural Plant Journal, 2023. doi: 10.1016/j.hpj.2023.01.004.
- Kong X#, Zhang Y#, Wang Z#, Bao S#, Feng Y, Wang J, Yu Z, Long F, Xiao Z, Hao Y, Gao X, Li Y, Ding Y, Wang J, Lei T, Xu C, Jinpeng Wang*, Two-step model of paleohexaploidy, ancestral genome reshuffling and plasticity of heat shock response in Asteraceae, Horticulture Research, 2023, 10(6):uhad073.
- Wenqian Kong#, Min Liu, Peter Felker, Mauricio Ewens, Cecilia Bessega, Carolina Pometti, Jinpeng Wang, Peng Xu, Jia Teng, Jinyu Wang, Xiyin Wang, Yuannian Jiao, Magdy S. Alabady, Françoise Thibaud-Nissen, Patrick Masterson, Xin Qiao, Andrew H. Paterson*, Genome and evolution of Prosopis alba Griseb., a drought and salinity tolerant tree legume crop for arid climates, Plants People Planet, 2023, 5:933-947.
- Jiaqi Wang#, Min Yuan#, Yishan Feng#, Yan Zhang#, Shoutong Bao, Yanan Hao, Yue Ding, Xintong Gao, Zijian Yu, Qiang Xu, Junxin Zhao, Qianwen Zhu, Ping Wang, Chunyang Wu, Jianyu Wang, Yuxian Li, Chuanyuan Xu, Jinpeng Wang*, A common whole-genome paleotetraploidization in Cucurbitales, Plant Physiology, Volume 190, Issue 4, December 2022, Pages 2430–2448.
- Jianyu Wang#, Ziyi Yang#, Tianyu Lei#, Yan Zhang, Qimeng Xiao, Zijian Yu, Jiaqi Zhang, Sangrong Sun, Qiang Xu, Shaoqi Shen, Zimo Yan, Mengnan Fang, Yue Ding, Zihan Liu, Qianwen Zhu, Ke Ren, Yuxin Pan*, Haibin Liu*, Jinpeng Wang*, A likely autotetraploidization event shaped the Chinese mahogany (Toona sinensis) genome, Horticultural Plant Journal, 2022, ISSN 2468-0141. Horticultural Plant Journal.2022.11.002.
- Yan Zhang#, Lan Zhang#, Qimeng Xiao#, Chunyang Wu#, Jiaqi Zhang, Qiang Xu, Zijian Yu, Shoutong Bao, Jianyu Wang, Yu Li, Li Wang*, Jinpeng Wang*. Two independent allohexaploidizations and genomic fractionation in Solanales. Front Plant Sci. 2022 Sep 23;13:1001402. PMID: 36212355; PMCID: PMC9538396.
- Jianyu Wang#, Lan Zhang#, Jiaqi Wang#, Yanan Hao, Qimeng Xiao, Jia Teng, Shaoqi Shen, Yan Zhang, Yishan Feng, Shoutong Bao, Yu Li, Zimo Yan, Chendan Wei, Li Wang, Jinpeng Wang* ,Conversion between duplicated genes generated by polyploidization contributes to the divergence of poplar and willow, BMC Plant Biol. 2022 Jun 17;22(1)
- Chunliu Zuo#, Lan Zhang#, Xinyue Yan#, Xinyue Guo, Qing Zhang, Songyang Li, Yanling Li, Wen Xu, Xiaoming Song, Jinpeng Wang*, Min Yuan*. Evolutionary analysis and functional characterization of BZR1 gene family in celery revealed their conserved roles in brassinosteroid signaling, BMC Genomics. 2022 Aug.
- Jia Teng#, Jianyu Wang, Lan Zhang, Chendan Wei, Shaoqi Shen, Qimeng Xiao, Yuanshuai Yue, Yanan Hao, Weina Ge, Jinpeng Wang*, Paleopolyploidies and Genomic Fractionation in Major Eudicot Clades, Front Plant Sci, 2022 May 31.
- Jinpeng Wang#, Pengchuan Sun, Yuxian Li, Yinzhe Liu, Nanshan Yang, Jigao Yu, Xuelian Ma, Sangrong Sun, Ruiyan Xia, Xiaojian Liu, Dongcen Ge, Sainan Luo, Yinmeng Liu, Youting Kong, Xiaobo Cui, Tianyu Lei, Li Wang, Zhenyi Wang, Weina Ge, Lan Zhang, Xiaoming Song, Min Yuan, Di Guo, Dianchuan Jin, Wei Chen, Yuxin Pan, Tao Liu, Guixian Yang, Yue Xiao, Jinshuai Sun, Cong Zhang, Zhibo Li, Haiqing Xu, Xueqian Duan, Shaoqi Shen, Zhonghua Zhang, Sanwen Huang,Xiyin Wang*, An overlooked paleo-tetraploidization in Cucurbitaceae,Molecular Biology and Evolution, 2018, 35(1): 16-26.
- Jinpeng Wang#, Pengchuan Sun, Yuxian Li, Yinzhe Liu, Jigao Yu, Xuelian Ma, Sangrong Sun, Nanshan Yang, Ruiyan Xia, Tianyu Lei, Xiaojian Liu, Beibei Jiao, Yue Xing, Weina Ge, Li Wang, Zhenyi Wang, Xiaoming Song, Min Yuan, Di Guo, Lan Zhang, Jiaqi Zhang, Dianchuan Jin, Wei Chen, Yuxin Pan, Tao Liu, Ling Jin, Jinshuai Sun, Jiaxiang Yu, Rui Cheng, Xueqian Duan, Shaoqi Shen, Jun Qin, Meng-chen Zhang, Andrew H. Paterson, Xiyin Wang*, Hierarchically aligning 10 legume genomes establishes a family-level genomics platform, Plant Physiology, 2017, 174 (1): 284-300.
- Jin-Peng Wang#, Ji-Gao Yu#, Jing Li#, Peng-Chuan Sun#, Li Wang#, Jia-Qing Yuan, Fan-Bo Meng, Sang-Rong Sun, Yu-Xian Li, Tian-Yu Lei, Yu-Xin Pan, Wei-Na Ge, Zhen-Yi Wang, Lan Zhang, Xiao-Ming Song, Chao Liu, Xue-Qian Duan, Shao-Qi Shen, Yang-qin Xie, Yue Hou, Jin Zhang, Jian-Yu Wang, Xiyin Wang*, Two likely auto-tetraploidization events shaped kiwifruit genome and contributed to establishment of the Actinidiaceae family, iScience, 2018, 7, 230-240.
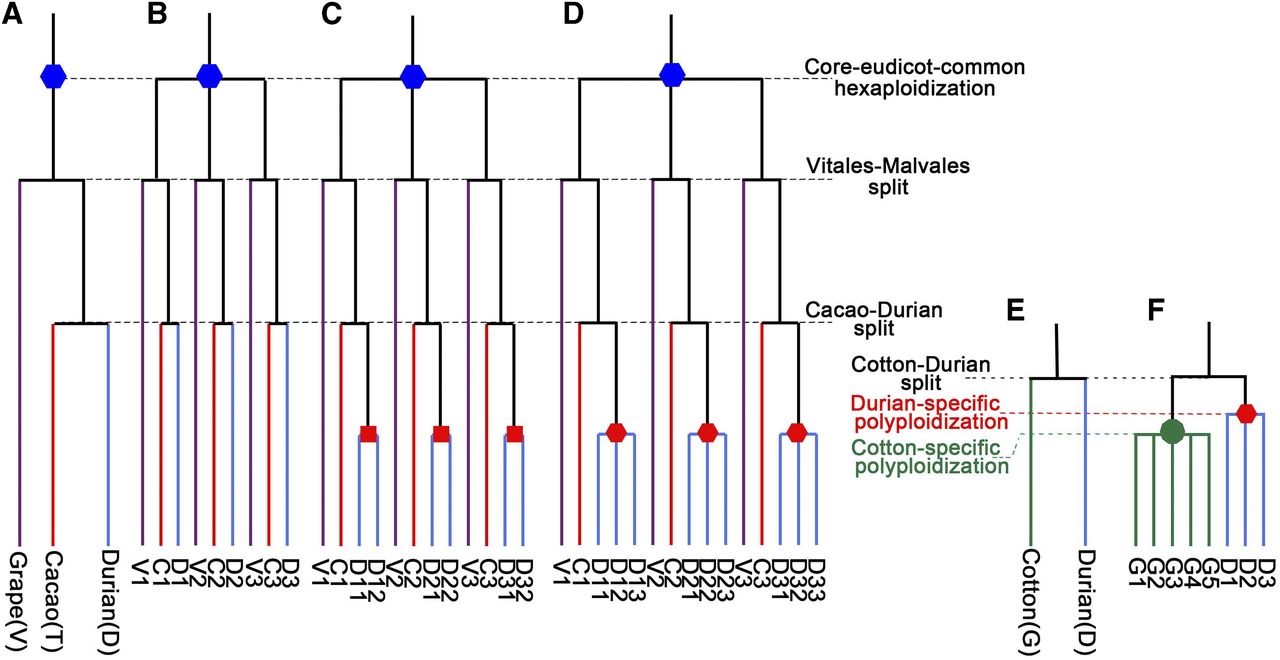
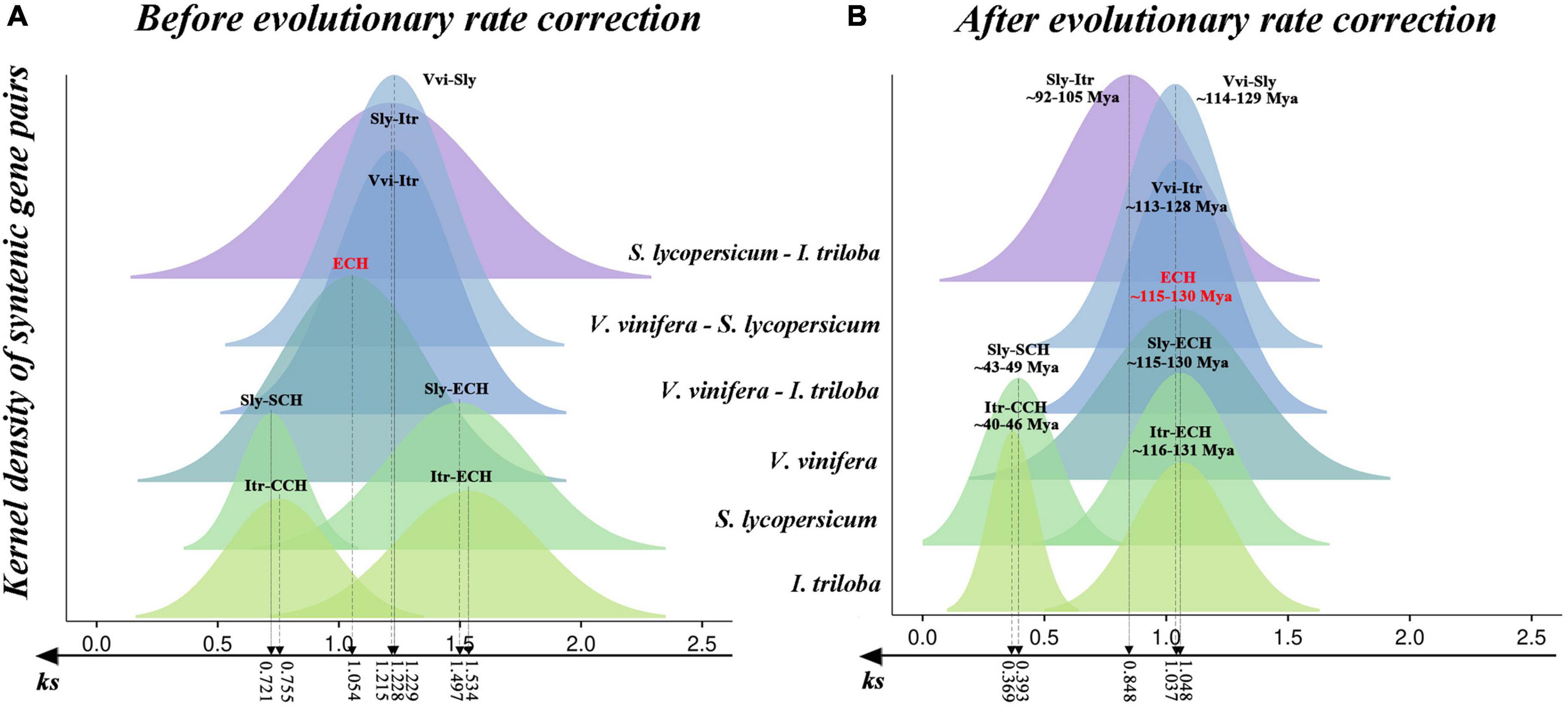
- Jinpeng Wang#, Jiaqing Yuan, Jigao Yu, Fanbo Meng, Pengchuan Sun, Yuxian Li, Nanshan Yang, Zhenyi Wang, Yuxin Pan, Weina Ge, Li Wang, Jing Li, Chao Liu, Zhiyan Xi, Yuhao Zhao, Sainan Luo, Dongcen Ge, Xiaobo Cui, Guangdong Feng, Ziwei Wang, Lei Ji, Jun Qin, Xiuqing Li, Xiyin Wang*, Recursive paleohexaploidization shapes the durian genome, Plant Physiology, 2019, 179 (1): 209-219.
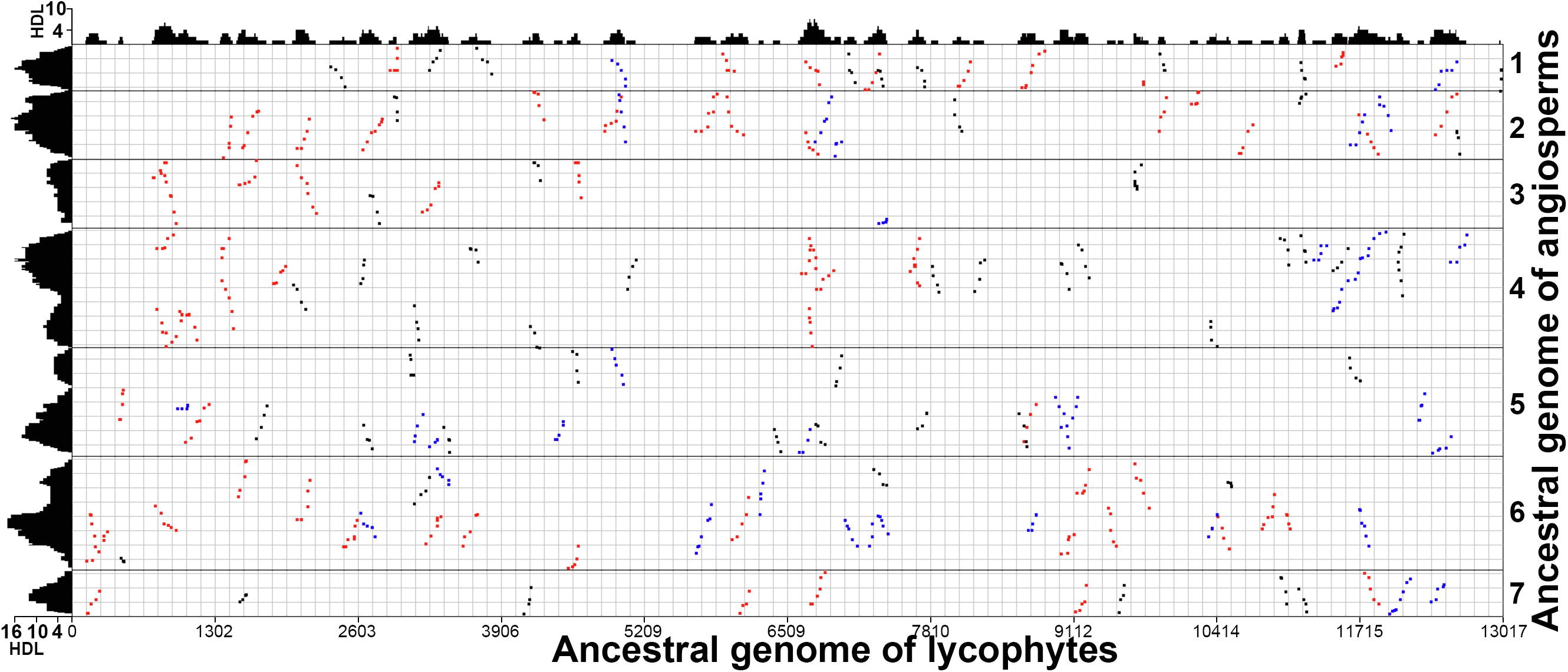
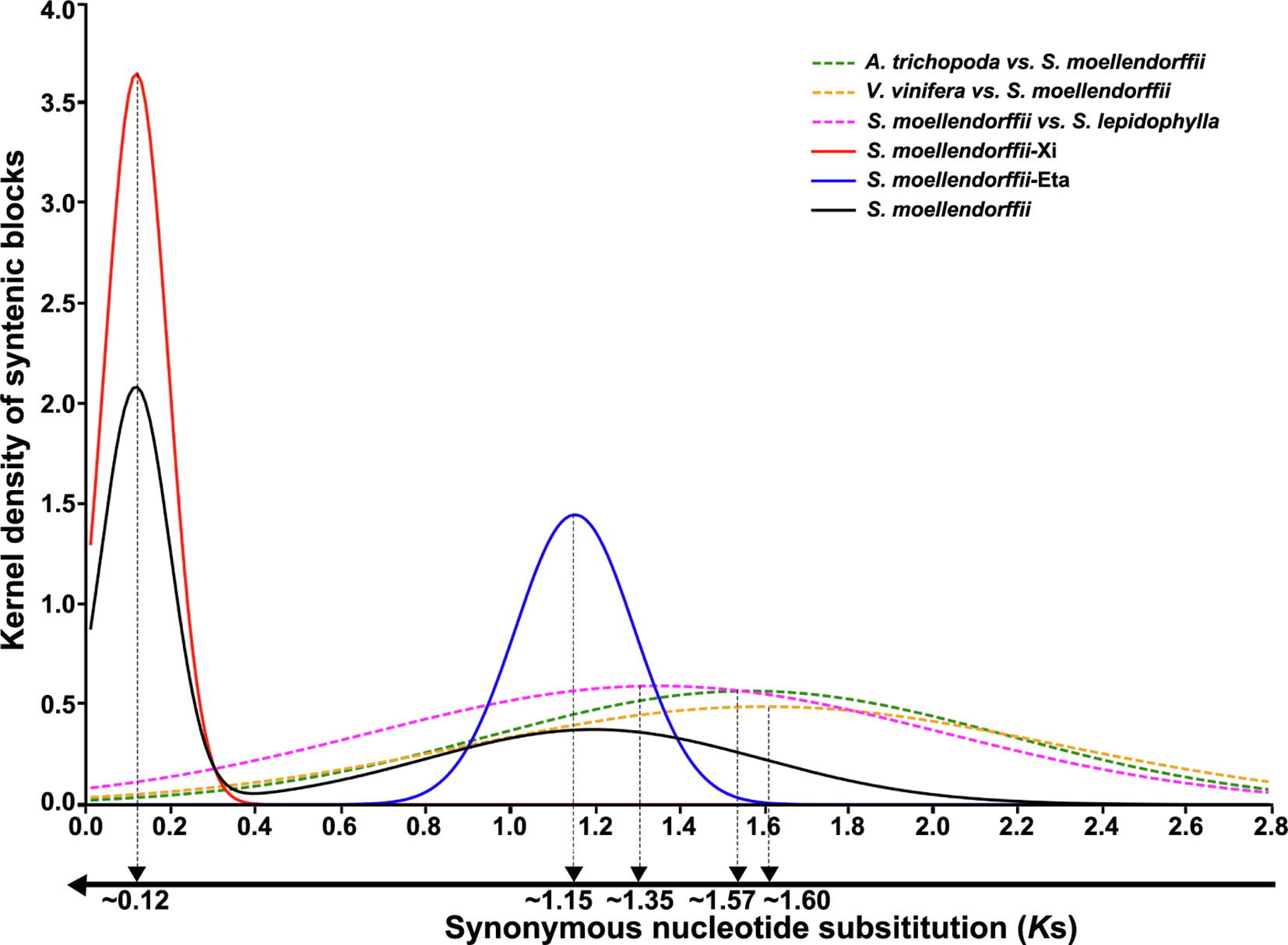
- Jinpeng Wang#, Jigao Yu, Pengchuan Sun, Chao Li, Xiaoming Song, Tianyu Lei, Yuxian Li, Jiaqing Yuan, Sangrong Sun, Hongling Ding, Xueqian Duan, Shaoqi Shen, Yanshuang Shen, Jing Li, Fanbo Meng, Yangqin Xie, Jianyu Wang, Yue Hou, Jin Zhang, Xianchun Zhang, Xiuqing Li, Andrew H. Paterson, Xiyin Wang*, Paleo-polyploidizations in lycophytes, Genomics Proteomics & Bioinformatics, 2019, 18 (3): 333-340.
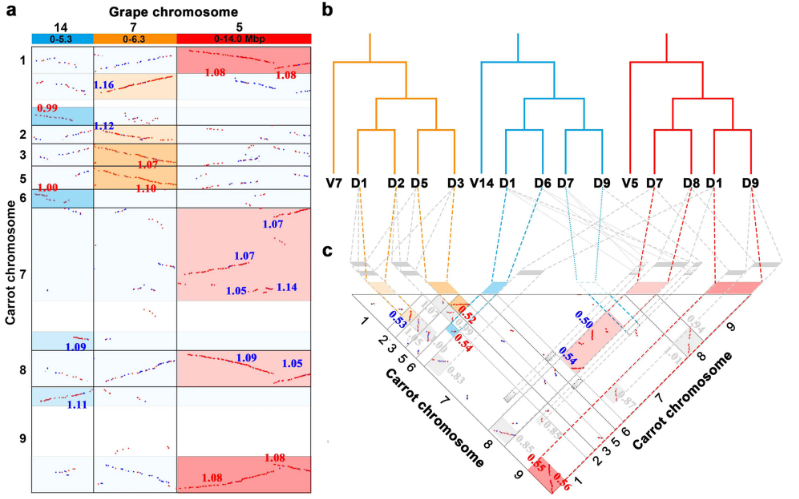
- Jinpeng Wang#, Jigao Yu#, Yuxian Li, Chendan Wei, He Guo, Ying Liu, Jin Zhang, Xiuqing Li, Xiyin Wang*. Sequential paleotetraploidization shaped the carrot genome, BMC plant biology, 2020, 20 (1): 52
- Chendan Wei#, Zhenyi Wang#, Jianyu Wang#, Jia Teng, Shaoqi Shen, Qimeng Xiao, Shoutong Bao, Yishan Feng, Yan Zhang, Yuxian Li, Sangrong Sun, Yuanshuai Yue, Chunyang Wu, Yanli Wang, Tianning Zhou, Wenbo Xu, Jigao Yu, Li Wang, Jinpeng Wang*. Conversion between 100-million-year-old duplicated genes contributes to rice subspecies divergence, BMC genomics, 2021, 22: 460
- Jinpeng Wang#, Jun Qin#, Pengchuan Sun, Xuelian Ma, Jigao Yu, Yuxian Li, Sangrong Sun, Tianyu Lei, Fanbo Meng, Chendan Wei, Xinyu Li, He Guo, Xiaojian Liu, Ruiyan Xia, Ruyue Zhang, Lei Ji, Li Wang, Weina Ge, Zhenyi Wang, Xiaoming Song, Lan Zhang, Di Guo, Yuxin Pan, Jinyu Wang, Shoutong Bao, Shan Jiang, Yishan Feng, Xueping Li, Andrew H. Paterson, Xiyin Wang*, Polyploidy index and its implications for the evolution of polyploids,Frontiers in genetics, 2019.
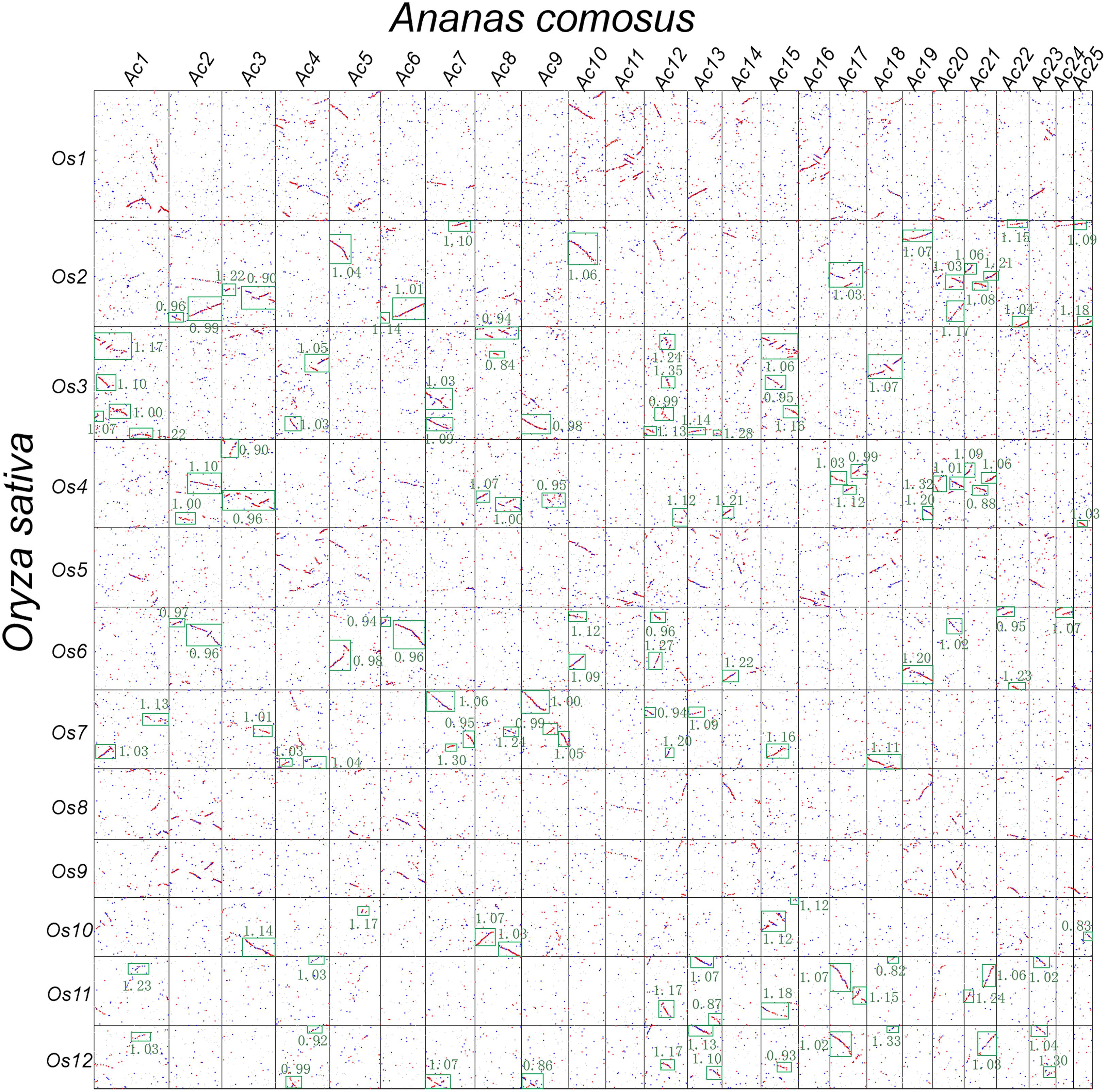
- Jinpeng Wang#, Jiaxiang Yu#, Pengchuan Sun, Yuxian Li, Ruiyan Xia, Yinzhe Liu, Xuelian Ma, Jigao Yu, Nanshan Yang, Tianyu Lei, Zhenyi Wang, Li Wang, Weina Ge, Xiaoming Song, Xiaojian Liu, Sangrong Sun, Tao Liu, Dianchuan Jin, Yuxin Pan*, Xiyin Wang*, Comparative genomics analysis of rice and pineapple contributes to understand the chromosome number reduction and genomic changes in grasses, Frontiers in Genetics, 2016, 10.3389 /fgene. 2016. 00174.
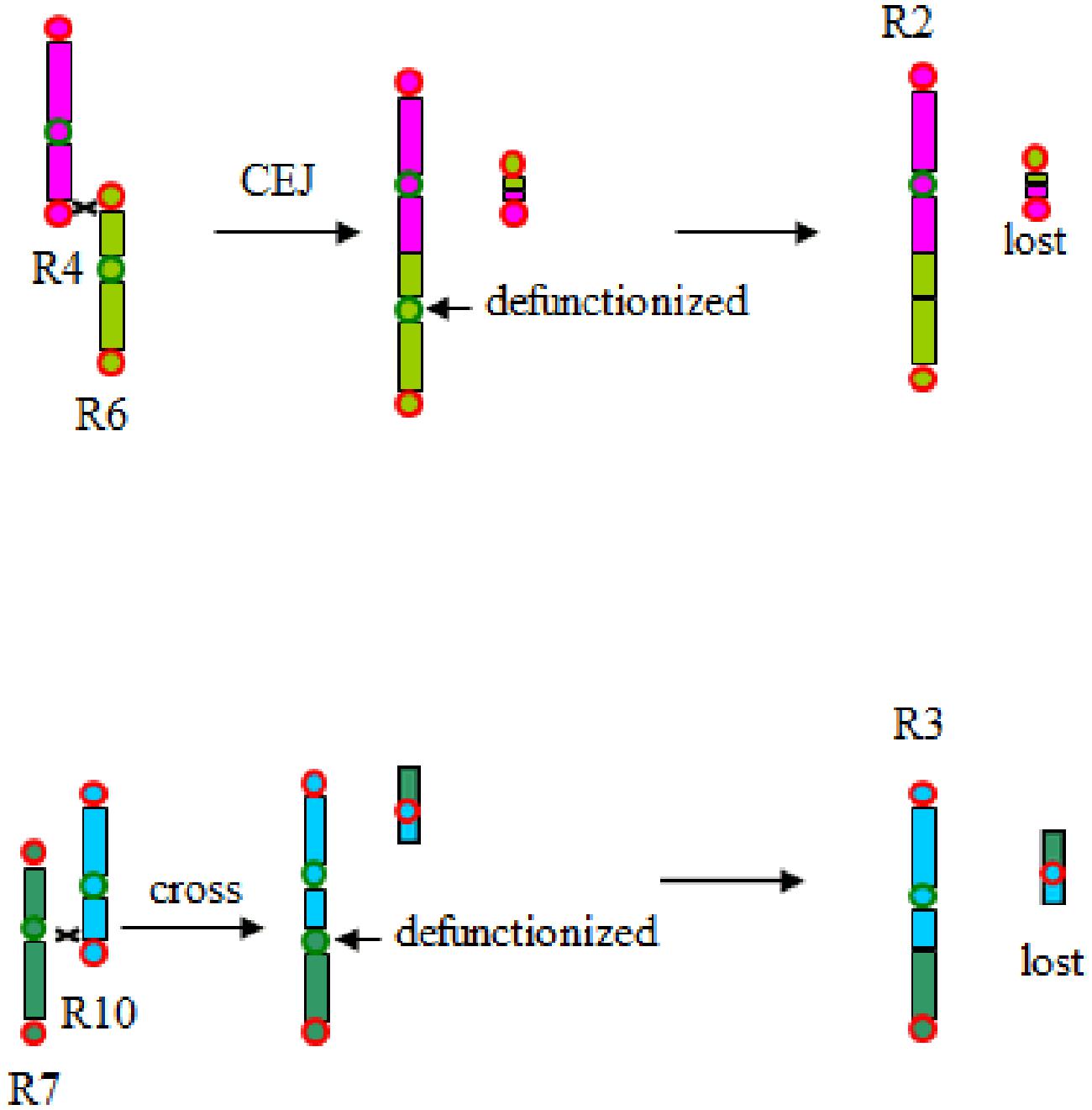
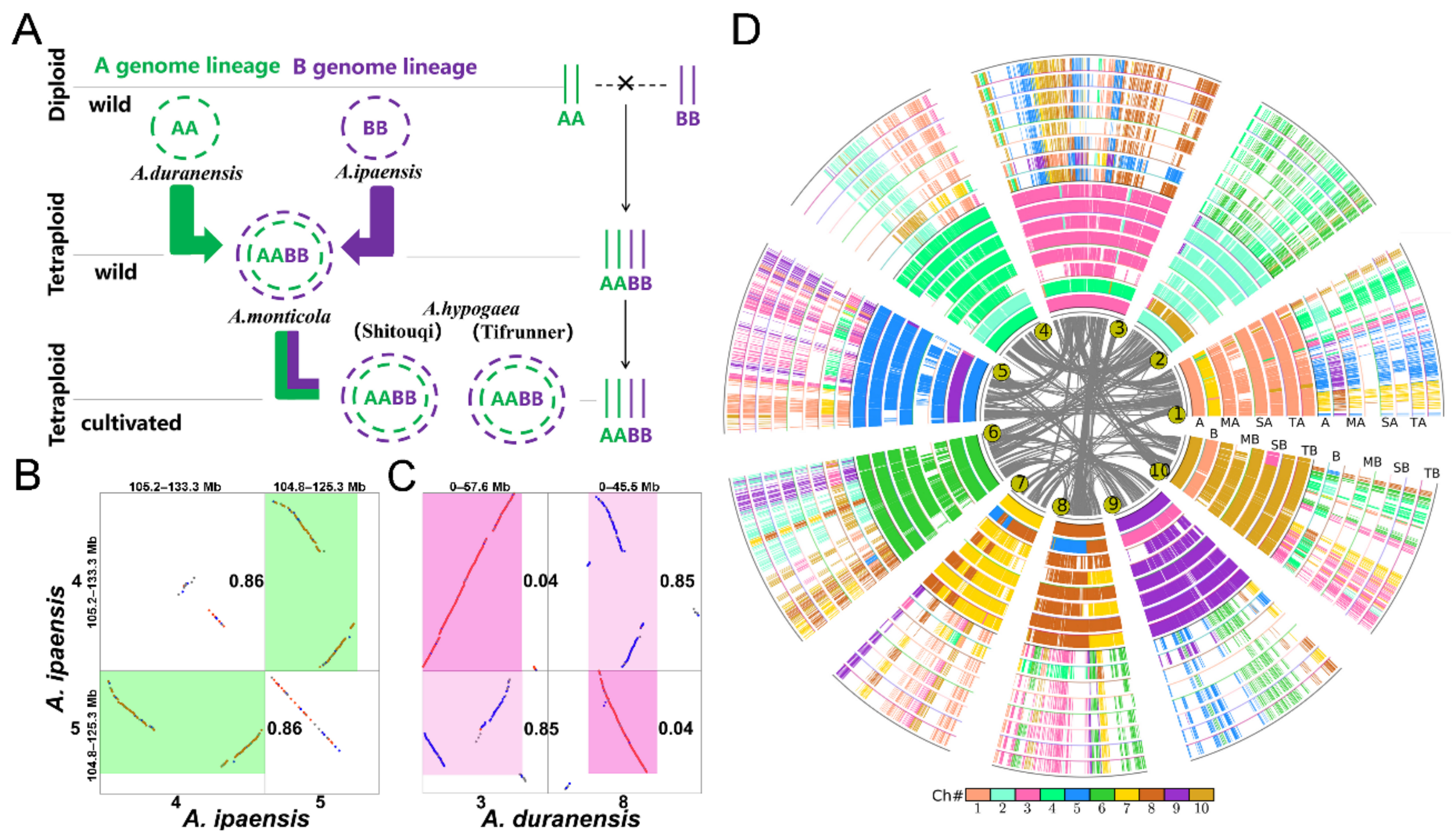
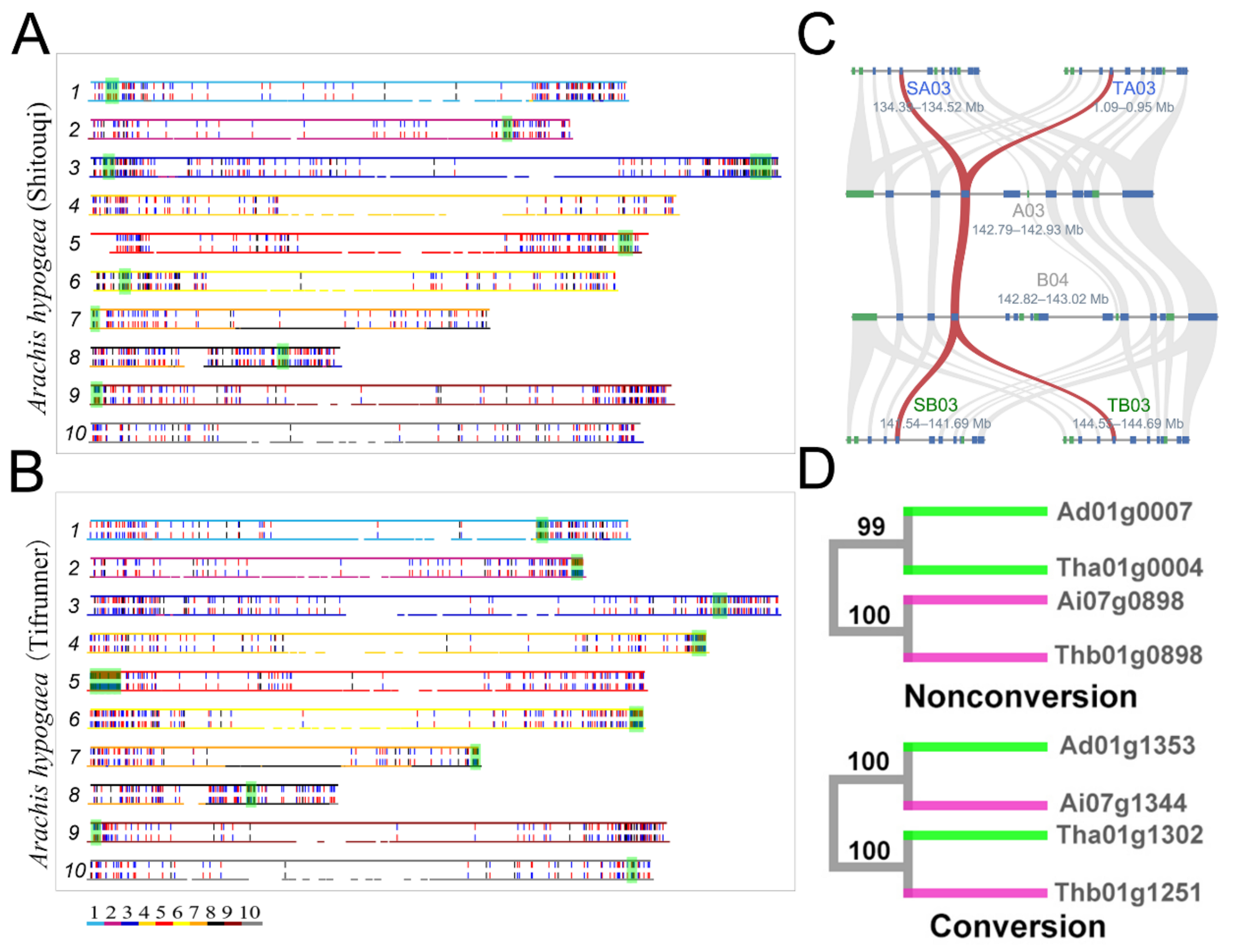
- Shaoqi Shen#, Yuxian Li#, Jianyu Wang, Chendan Wei, Zhenyi Wang, Weina Ge, Min Yuan, Lan Zhang, Li Wang, Sangrong Sun, Jia Teng, Qimeng Xiao, Shoutong Bao, Yishan Feng, Yan Zhang, Jiaqi Wang, Yanan Hao, Tianyu Lei*,Jinpeng Wang*. Illegitimate Recombination between Duplicated Genes Generated from Recursive Polyploidizations Accelerated the Divergence of the Genus Arachis, Genes, 2021, 12, 1944.
- Liuyu Qin#, Yiheng Hu#, Jinpeng Wang#, Xiaoliang Wang#, Ran Zhao#, Hongyan Shan, Kunpeng Li, Peng Xu , Hanying Wu, Xueqing Yan, Lumei Liu, Xin Yi, Stefan Wanke , John E. Bowers, James H. Leebens-Mack, Claude W. dePamphilis , Pamela S. Soltis , Douglas E. Soltis , Hongzhi Kong , Yuannian Jiao *. Insights into angiosperm evolution, floral development and chemical biosynthesis from the Aristolochia fimbriata genome, Nature Plants, 2021
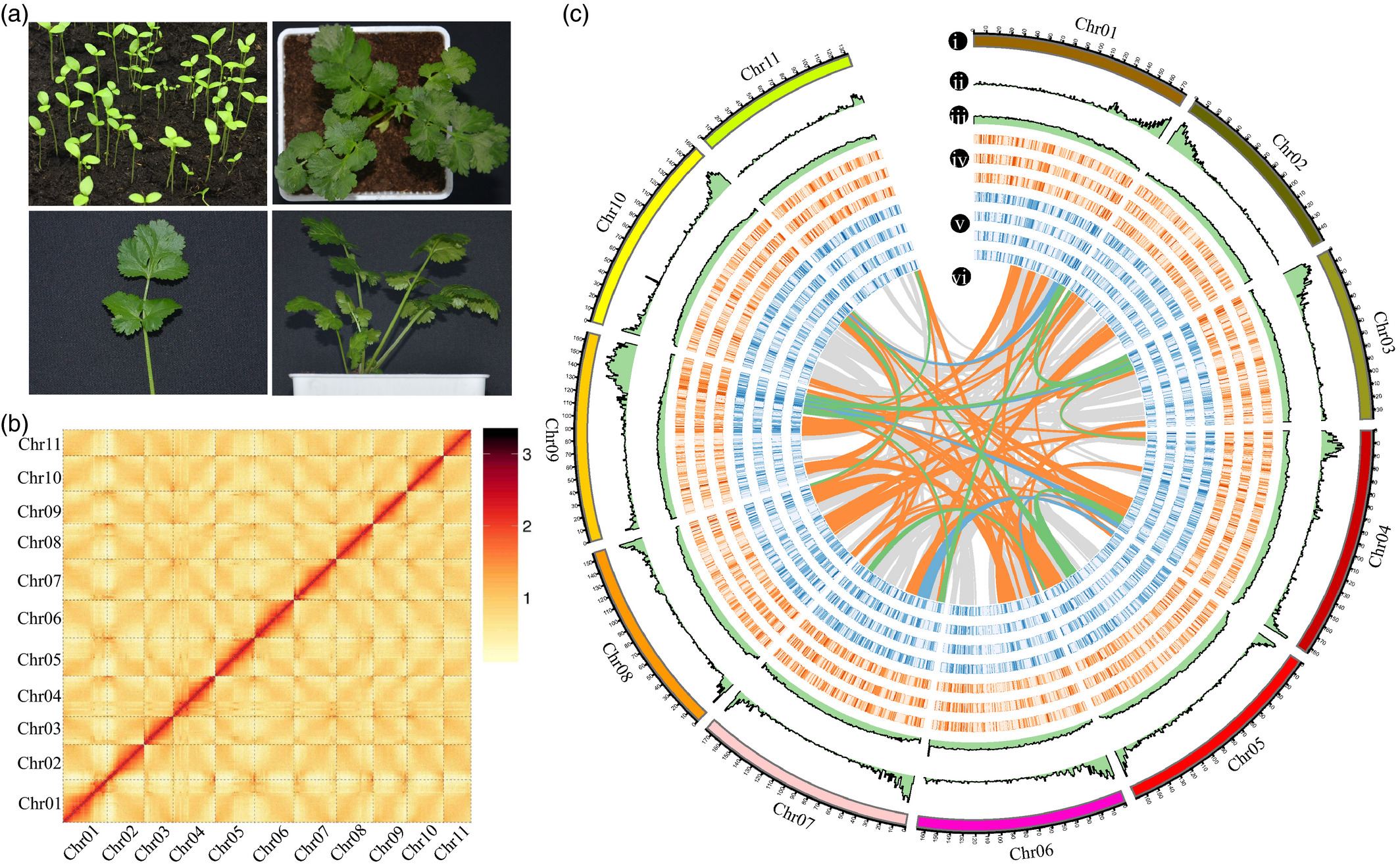
- Xiaoming Song#, Jinpeng Wang#, Nan Li, Jigao Yu, Fanbo Meng, Chendan Wei, Chao Liu,Wei Chen, Fulei Nie, Zhikang Zhang, Ke Gong, Xinyu Li, Jingjing Hu, Qihang Yang, Yuxian Li,Chunjin Li, Shuyan Feng, He Guo, Jiaqing Yuan, Qiaoying Pei, Tong Yu, Xi Kang, Wei Zhao, Tianyu Lei,Pengchuan Sun, Li Wang, Weina Ge, Di Guo, Xueqian Duan, Shaoqi Shen, Chunlin Cui, Ying Yu,Yangqin Xie, Jin Zhang, Yue Hou, Jianyu Wang, Jinyu Wang, Xiu-Qing Li, Andrew H. Paterson, Xiyin Wang*. Deciphering the high-quality genome sequence of coriander that causes controversial feelings, 2019, doi:https://onlinelibrary.wiley.com/doi/10.1111/pbi.13310
- Yinzhe Liu#, Jinpeng Wang#, Weina Ge, Zhenyi Wang, Yuxian Li, Nanshan Yang, Sangrong Sun, Liwei Zhang, Xiyin Wang*, Two Highly Similar Poplar Paleo-subgenomes Suggest an Autotetraploid Ancestor of Salicaceae Plants,Front Plant Sci, 2017, 8: 571.
- Sangrong Sun#, Jinpeng Wang#, Jigao Yu, Fanbo Meng, Ruiyan Xia, Li Wang, Zhenyi Wang, Weina Ge, Xiaojian Liu, Yuxian Li, Yinzhe Liu, Nanshan Yang, Xiyin Wang*, Alignment of Common Wheat and Other Grass Genomes Establishes a Comparative Genomics Research Platform, Front Plant Sci, 2017, 8: 1480.
- Jiaqing Yuan#, Jinpeng Wang#, Jigao Yu, Fanbo Meng, Yuhao Zhao, Jing Li, Pengchuan Sun, Sangrong Sun, Zhikang Zhang, Chao Liu, Chendan Wei, He Guo, Xinyu Li, Xueqian Duan, Shaoqi Shen, Yangqin Xie, Yue Hou, Jin Zhang, Tariq Shehzad* , Xiyin Wang*. Alignment of Rutaceae Genomes Reveals Lower Genome Fractionation Level Than Eudicot Genomes Affected by Extra Polyploidization, Frontiers in plant Science, 2019, 10:986. doi: 10.3389/fpls.2019.00986.
- Zhenyi Wang#, Jinpeng Wang#, Yuxin Pan, Tianyu Lei, Weina Ge, Li Wang, Lan Zhang, Yuxian Li, Kanglu Zhao, Tao Liu, Xiaoming Song, Jiaqi Zhang, Jigao Yu, Jingjing Hu, Xiyin Wang*. Reconstruction of evolutionary trajectories of chromosomes unraveled independent genomic repatterning between Triticeae and Brachypodium, 2019, https://doi.org/10.1186/s12864-019-5566-8.
- Vanika Garg#, Olga Dudchenko#, Jinpeng Wang#, Aamir W. Khan, Saurabh Gupta, Parwinder Kaur, Kai Han, Rachit K. Saxena, Sandip M. Kale, Melanie Pham, Jigao Yu, Annapurna Chitikineni, Zhikang Zhang, Guangyi Fan, Christopher Lui, Vinodkumar Valluri, Fanbo Meng, Aditi Bhandari, Xiaochuan Liu, Tao Yang, Hua Chen, Babu Valliyodan, Manish Roorkiwal, Chengcheng Shi, Hong Bin Yang, Neva C. Durand, Manish K. Pandey, Guowei Li, Rutwik Barmukh, Xingjun Wang, Xiaoping Chen, Hon-Ming Lam, Huifang Jiang, Xuxiao Zong, Xuanqiang Liang, Xin Liu, Boshou Liao, Baozhu Guo, Scott Jackson, Henry T. Nguyen, Weijian Zhuang, Wan Shubo*, Xiyin Wang*, Erez Lieberman Aiden*, Jeffrey L. Bennetzen*, Rajeev K. Varshney*. Chromosome-length genome assemblies of six legume species provide insights into genome organization, evolution, and agronomic traits for crop improvement, Journal of Advanced Research, 2021
- Fanbo Meng#, Yuxin Pan#, Jinpeng Wang#, Jigao Yu, Chao Liu, Zhikang Zhang, Chendan Wei, He Guo, Xiyin Wang*. Cotton Duplicated Genes Produced by Polyploidy Show Significantly Elevated and Unbalanced Evolutionary Rates, Overwhelmingly Perturbing Gene Tree Topology, 2020, 11(239), doi: https://www.frontiersin.org/articles/10.3389/fgene.2022.902804/full.
- Zhenyi Wang#, Kanglu Zhao#, Yuxin Pan#, Jinpeng Wang#, Xiaoming Song#, Weina Ge#, Min Yuan, Tianyu Lei, Li Wang, Lan Zhang, Yuxian Li, Tao Liu, Wei Chen, Wenjing Meng, Changkai Sun, Xiaobo Cui, Yun Bai, Xiyin Wang*, Genomic, expressional, protein-protein interactional analysis of Trihelix transcription factor genes in Setaria italia and inference of their evolutionary trajectory BMC Genomics, 2018, .
- Jiale Yuan#, Ying Liu#, Zhenyi Wang, Tianyu Lei, Yanfang Hu, Lan Zhang, Min Yuan, Jinpeng Wang*, Yuxian Li*. Genome-Wide Analysis of the NAC Family Associated with Two Paleohexaploidization Events in the Tomato, Life, 2022.
- Jia Teng#, Jianyu Wang#, Lan Zhang#, Chendan Wei, Shaoqi Shen, Qimeng Xiao, Yuanshuai Yue, Yanan Hao, Weina Ge*, Jinpeng Wang*. Paleopolyploidies and Genomic Fractionation in Major Eudicot Clades, Front Plant Sci, 2022 May 31.
- Chunliu Zuo#, Lan Zhang#, Xinyue Yan#, Xinyue Guo, Qing Zhang, Songyang Li, Yanling Li, Wen Xu, Xiaoming Song , Jinpeng Wang* , Min Yuan*. Evolutionary analysis and functional characterization of BZR1 gene family in celery revealed their conserved roles in brassinosteroid signaling, BMC Genomics. 2022 Aug.
- Jianyu Wang#, Lan Zhang#, Jiaqi Wang#, Yanan Hao, Qimeng Xiao, Jia Teng, Shaoqi Shen, Yan Zhang, Yishan Feng, Shoutong Bao, Yu Li, Zimo Yan, Chendan Wei, Li Wang*, Jinpeng Wang*. Conversion between duplicated genes generated by polyploidization contributes to the divergence of poplar and willow, BMC Plant Biol. 2022 Jun 17;22(1).
- Jiaqi Wang#, Min Yuan#, Yishan Feng#, Yan Zhang, Shoutong Bao, Yanan Hao, Yue Ding, Xintong Gao, Zijian Yu, Qiang Xu, Junxin Zhao, Qianwen Zhu, Ping Wang, Chunyang Wu, Jianyu Wang, Yuxian Li, Chuanyuan Xu, Jinpeng Wang*, A common whole-genome paleotetraploidization in Cucurbitales, Plant Physiology, Volume 190, Issue 4, December 2022, Pages 2430–2448.
- Yan Zhang#, Lan Zhang#, Qimeng Xiao#, Chunyang Wu#, Jiaqi Zhang, Qiang Xu, Zijian Yu, Shoutong Bao, Jianyu Wang, Yu Li, Li Wang*, Jinpeng Wang*. Two independent allohexaploidizations and genomic fractionation in Solanales. Front Plant Sci
- Jianyu Wang#, Ziyi Yang#, Tianyu Lei#, Yan Zhang, Qimeng Xiao, Zijian Yu, Jiaqi Zhang, Sangrong Sun, Qiang Xu, Shaoqi Shen, Zimo Yan, Mengnan Fang, Yue Ding, Zihan Liu, Qianwen Zhu, Ke Ren, Yuxin Pan*, Haibin Liu*, Jinpeng Wang*. A likely autotetraploidization event shaped the Chinese mahogany (Toona sinensis) genome, Horticultural Plant Journal, 2022, ISSN 2468-0141.
- Zhi-Kun Li#, Bin Chen#, Xiu-Xin Li, Jin-Peng Wang, Yan Zhang, Xing-Fen Wang, Yuan-Yuan Yan, Hui-Feng Ke,Jun Yang, Jin-Hua Wu, Guo-Ning Wang, Gui-Yin Zhang, Li-Qiang Wu, Xi-Yin Wang*, Zhi-Ying Ma*. A newly identified cluster of glutathione S-transferase genesprovides Verticillium wilt resistance in cotton, The plant Journal, 2019, 98, 213–227.
- Xiaoping Chen#, Qing Lu#, Hao Liu#, Jianan Zhang#, Yanbin Hong#, Haofa Lan, Haifen Li, Jinpeng Wang, Haiyan Liu, Shaoxiong Li, Manish K. Pandey, Zhikang Zhang, Guiyuan Zhou, Jigao Yu, Guoqiang Zhang, Jiaqing Yuan, Xingyu Li, Shijie Wen, Fanbo Meng, Shanlin Yu, Xiyin Wang, Kadambot H M Siddique, Zhong-Jian Liu*, Andrew H. Paterson*, Rajeev K. Varshney*, Xuanqiang Liang*. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement, Molecular Plant, 2019, 2018-21070.
- Xiaoming Song#, Jinpeng Wang, Xiao Ma, Yuxian Li, Tianyu Lei, Li Wang, Weina Ge, Di Guo, Zhenyi Wang, Chunjin Li, Jianjun Zhao*, Xiyin Wang*. Origination, Expansion, Evolutionary Trajectory, and Expression Bias of AP2/ERF Superfamily in Brassica napus, Front Plant Sci, 2016, 7: 1186.
- Xiyin Wang#, Hui Guo#, Jinpeng Wang#, Tianyu Lei, Tao Liu, Zhenyi Wang, Yuxian Li, Tae-Ho Lee, Jingping Li, Haibao Tang, Dianchuan Jin, Andrew H Paterson*. Comparative genomic de-convolution of the cotton genome revealed a decaploid ancestor and widespread chromosomal fractionation, New Phytologist, 2016, 209(3): 1252-1263.
- Xiyin Wang#, Jinpeng Wang, Dianchuan Jin, Hui Guo, Tae-Ho Lee, Tao Liu, Andrew H. Paterson*. Genome alignment spanning major Poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key events, Molecular Plant, 2015, 8(6), 885-898.
- Shengyi Liu#, Yumei Liu#, Xinhua Yang#, Chaobo Tong#, David Edwards#, Isobel A P Parkin#, Meixia Zhao, Jianxin Ma, Jingyin Yu, Shunmou Huang, Xiyin Wang, Junyi Wang, Kun Lu, Zhiyuan Fang, Ian Bancroft, Tae-Jin Yang, Qiong Hu, Xinfa Wang, Zhen Yue, Haojie Li, Linfeng Yang, Jian Wu, Qing Zhou, Wanxin Wang, Graham J King, J Chris Pires, Changxin Lu, Zhangyan Wu, Perumal Sampath, Zhuo Wang, Hui Guo, Shengkai Pan, Limei Yang, Jiumeng Min, Dong Zhang, Dianchuan Jin, Wanshun Li, Harry Belcram, Jinxing Tu, Mei Guan, Cunkou Qi, Dezhi Du, Jiana Li, Liangcai Jiang, Jacqueline Batley, Andrew G Sharpe, Beom-Seok Park, Pradeep Ruperao, Feng Cheng, Nomar Espinosa Waminal, Yin Huang, Caihua Dong, Li Wang, Jingping Li, Zhiyong Hu, Mu Zhuang, Yi Huang, Junyan Huang, Jiaqin Shi, Desheng Mei, Jing Liu, Tae-Ho Lee, Jinpeng Wang, Huizhe Jin, Zaiyun Li, Xun Li, Jiefu Zhang, Lu Xiao, Yongming Zhou, Zhongsong Liu, Xuequn Liu, Rui Qin, Xu Tang, Wenbin Liu, Yupeng Wang, Yangyong Zhang, Jonghoon Lee, Hyun Hee Kim, France Denoeud, Xun Xu, Xinming Liang, Wei Hua, Xiaowu Wang, Jun Wang, Boulos Chalhoub, Andrew H Paterson*. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes, Nature Communications, 2014, 5: 3930.
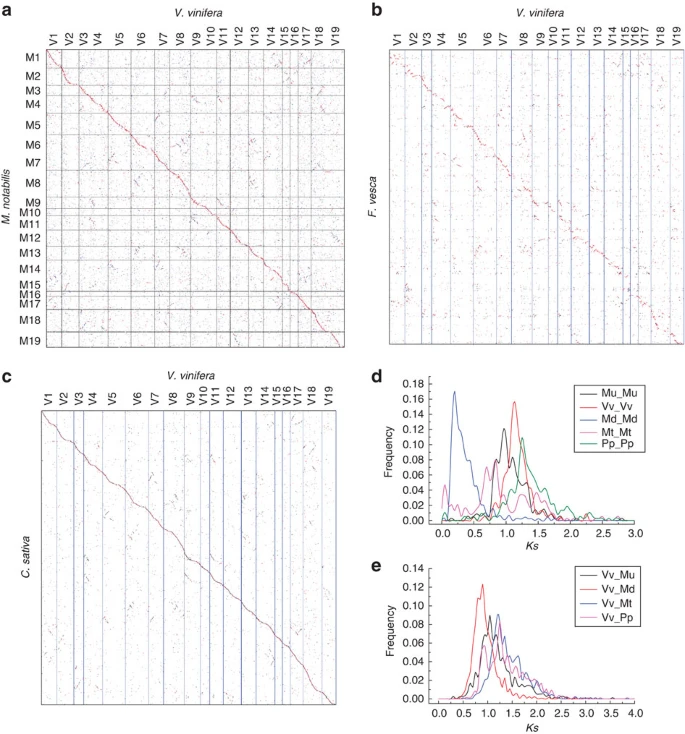
- Ningjia He#, Chi Zhang, Xiwu Qi, Shancen Zhao, Yong Tao, Guojun Yang, Tae-Ho Lee, Xiyin Wang, Qingle Cai, Dong Li, Mengzhu Lu, Sentai Liao, Guoqing Luo, Rongjun He, Xu Tan, Yunmin Xu, Tian Li, Aichun Zhao, Ling Jia, Qiang Fu, Qiwei Zeng, Chuan Gao, Bi Ma, Jiubo Liang, Xiling Wang, Jingzhe Shang, Penghua Song, Haiyang Wu, Li Fan, Qing Wang, Qin Shuai, Juanjuan Zhu, Congjin Wei, Keyan Zhu-Salzman, Dianchuan Jin, Jinpeng Wang, Tao Liu, Maode Yu, Cuiming Tang, Zhenjiang Wang, Fanwei Dai, Jiafei Chen, Yan Liu, Shutang Zhao, Tianbao Lin, Shougong Zhang, Junyi Wang, Jian Wang, Huanming Yang, Guangwei Yang, Jun Wang*, Andrew H. Paterson, Qingyou Xia, Dongfeng Ji*, Zhonghuai Xiang*. Draft genome sequence of the mulberry tree Morus notabilis, Nature Communications, 2013, 4: 2445.
- Andrew H. Paterson#, Jonathan F. Wendel, Heidrun Gundlach, Hui Guo, Jerry Jenkins, Dianchuan Jin, Danny Llewellyn, Kurtis C. Showmaker, Shengqiang Shu, Joshua Udall, Mi-jeong Yoo, Robert Byers, Wei Chen, Adi Doron-Faigenboim, Mary V. Duke, Lei Gong, Jane Grimwood, Corrinne Grover, Kara Grupp, Guanjing Hu, Tae-ho Lee, Jingping Li, Lifeng Lin, Tao Liu, Barry S. Marler, Justin T. Page, Alison W. Roberts, Elisson Romanel, William S. Sanders, Emmanuel Szadkowski, Xu Tan, Haibao Tang, Chunming Xu, Jinpeng Wang, Zining Wang, Dong Zhang, Lan Zhang, Hamid Ashrafi, Frank Bedon, John E. Bowers, Curt L. Brubaker, Peng W. Chee, Sayan Das, Alan R. Gingle, Candace H. Haigler, David Harker, Lucia V. Hoffmann, Ran Hovav, Donald C. Jones, Cornelia Lemke, Shahid Mansoor, Mehboob ur Rahman, Lisa N. Rainville, Aditi Rambani, Umesh K. Reddy, Jun-kang Rong, Yehoshua Saranga, Brian E. Scheffler, Jodi A. Scheffler, David M. Stelly, Barbara A. Triplett, Allen Van Deynze, Maite F. S. Vaslin, Vijay N. Waghmare, Sally A. Walford, Robert J. Wright, Essam A. Zaki, Tianzhen Zhang, Elizabeth S. Dennis, Klaus F. X. Mayer, Daniel G. Peterson, Daniel S. Rokhsar, Xiyin Wang, Jeremy Schmutz*. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres, Nature, 2012, 492: 423-427.
- Xiaowu wang#, Hanzhong wang#, Jun wang#, Rifei Sun#, Jian Wu, Shengyi liu, Yinqi Bai, Jeong-Hwan mun, Ian Bancroft, Feng Cheng, Sanwen Huang, Xixiang Li, Wei Hua, Junyi wang, Xiyin wang, Michael Freeling, J Chris Pires, Andrew H Paterson, Boulos Chalhoub, Bo Alice Hayward, Andrew G Sharpe, Beom-Seok Park, Bernd weisshaar, Binghang liu, Bo Li, Bo liu, Chaobo Tong, Chi Song, Christopher duran, Chunfang Peng, Chunyu Geng, Chushin koh, Chuyu lin, david edwards, Desheng Mu, Di Shen, Eleni Soumpourou, Fei Fiona Fraser, Gavin Conant, Gilles lassalle, Graham J king, Guusje Bonnema, Haibao Tang, Haiping wang, Harry Belcram, Heling Zhou, Hideki Hirakawa, Hiroshi Abe, Hui Guo, Hui Huizhe Jin, Isobel A P Parkin, Jacqueline Batley, Jeong-Sun kim, Jérémy Just, Jianwen Li, Jiaohui Xu, Jie deng, Jin A Jingping Li, Jingyin Yu, Jinling meng, Jinpeng Wang, Jiumeng min, Julie Poulain, Jun wang, Katsunori Hatakeyama, Kui Wu, Li Lu Fang, Martin Trick, matthew G links, Meixia Zhao, Mina Jin, Nirala Ramchiary, nizar drou, Paul J Berkman, Qingle Cai, Quanfei Huang, Ruiqiang Satoshi Tabata, Shifeng Cheng, Shu Zhang, Shujiang Zhang, Shunmou Huang, Shusei Sato, Silong Sun, Soo-Jin kwon, Su-Ryun Choi, Tae-Ho lee, Wei Fan, Xiang Zhao, Xu Tan, Xun Xu, Yan wang, Yang Qiu, Ye Yin, Yingrui Li, Yongchen Du, Yongcui liao, Yongpyo lim, Yoshihiro narusaka, Yupeng wang, Zhenyi wang, Zhenyu Zhi, wen wang, Zhiyong Xiong, Zhonghua Zhang*. The genome of the mesopolyploid crop species Brassica rapa, Nature Genetics, 2011, 43: 1035-1039.
- Dasen Xie#, Yuanchao Xu#, Jinpeng Wang, Wenrui Liu, Qian Zhou, Shaobo Luo, Wu Huang, Xiaoming He, Qing Li, Qingwu Peng, Xueyong Yang, Jiaqing Yuan, Jigao Yu, Xiyin Wang, William J Lucas, Sanwen Huang, Biao Jiang*, Zhonghua Zhang*. The wax gourd genomes offer insights into the ancestral cucurbit karyotype and the genetic basis of diversity, Nature Communications, 2019.
- Weijian Zhuang#, Hua Chen#, Meng Yang#, Jianping Wang#, Manish K. Pandey, Chong Zhang, Wen-Chi Chang, Liangsheng Zhang, Xingtan Zhang, Ronghua Tang, Vanika Garg, Xingjun Wang, Haibao Tang, Chi-Nga Chow, Jinpeng Wang, Ye Deng , Depeng Wang, Aamir W. Khan, Qiang Yang, Tie-Cheng Cai, Prasad Bajaj, Kangcheng Wu, Baozhu Guo, Xinyou Zhang, Jingjing Li, Fan Liang, Jiang Hu, Boshou Liao, Shengyi Liu, Annapurna Chitikineni, Songhan Yan, Yixiong Zheng, Shihua Shan, Qinzheng Liu, Dongyang Xie, Zhenyi Wang, Shahid Ali Khan, Niaz Ali, Chuanzhi Zhao, Xinguo Li, Ziliang Luo, Shubiao Zhang, Rui-Rong Zhuang, Ze Peng, Shuai-Yin Wang, Gandeka Mamadou, Yuhui Zhuang, Zifan Zhao, Weichang Yu, Faqian Xiong, Weipeng Quan, Mei Yuan, Yu Li, Huashong Zou, Han Xia, Li Zha , Junpeng Fan, Jigao Yu, Wenping Xie, Jiaqing Yuan, Kun Chen, Shanshan Zhao, Wenting Chu, Yuting Chen, Pengchuan Sun, Fanbo Meng, Tao Zhuo, Yuhao Zhao, Chun-Juan Li, Guohao He, Yongli Zhao, Congcong Wang, Polavarapu Bilhan Kavikishor, Rong-Long Pan, Andrew H. Paterson, Xiyin Wang*, Ray Ming*, Rajeev K Varshney*. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication, Nature Genetics, 2019.
- Chao Liu#, Jinpeng Wang#, Pengchuan Sun, Jigao Yu, Fanbo Meng, Zhikang Zhang, He Guo, Chendan Wei, Xinyu Li, Shaoqi Shen, Xiyin Wang*. Illegitimate Recombination between Homeologous Genes in Wheat Genome, Frontiers in plant science, 2020.
- Jin Liu#, Cong Shi, Cheng-Cheng Shi, Wei Li, Qun-Jie Zhang, Yun Zhang, Kui Li, Hui-Fang Lu, Chao Shi, Si-Tao Zhu, Zai-Yun Xiao , Hong Nan, Yao Yue, Xun-Ge Zhu, Yu Wu, Xiao-Ning Hong, Guang-Yi Fan, Yan Tong, Dan Zhang, Chang-Li Mao, Yun-Long Liu, Shi-Jie Hao, Wei-Qing Liu, Mei-Qi Lv, Hai-Bin Zhang, Yuan Liu, Ge-Ran Hu-tang, Jin-peng Wang, Jia Hao Wang, Ying-Huai Sun, Shu-bang Ni, Wen-Bin Chen, Xing-Cai Zhang, Yuan-nian Jiao, Evan E. Eichler, Guo-hua Li, Xin Liu*, Li-Zhi Gao*. The Chromosome-based Rubber Tree Genome Provides New Insights into Spurge Genome Evolution and Rubber Biosynthesis, Molecular Plant, 2019.
- Xiao-Ming Song#, Jin-Peng Wang#, Peng-Chuan Sun, Xiao Ma, Qi-Hang Yang, Jing-Jing Hu, Sang-Rong Sun, Yu-Xian Li, Ji-Gao Yu, Shu-Yan Feng, Qiao-Ying Pei, Tong Yu, Nan-Shan Yang, Yin-Zhe Liu, Xiu-Qing Li, Andrew H. Paterson, Xi-Yin Wang*. Preferential gene retention increases the robustness of cold regulation in Brassicaceae and other plants after polyploidization, Horticulture Research, 2020.