Nov. 6, 2022 Allopolyploidizations and highly dynamic reshuffling of paleogenome contributed to the successful establishment of Orchidaceae
Nov. 15, 2022 Two-step model of paleohexaploidy, reshuffling of ancient genome and plasticity of heat shock response in Asteraceae
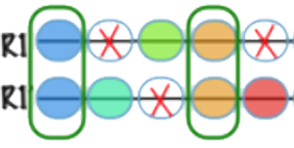
Nov. 23, 2022 ・Multi-dimensional reshuffling of ancestral genome during post-polyploid diploidization shaped family Fabaceae
2024 Aug 31 ・Transcriptome-wide identification of the Hsp70 gene family in Pugionium cornutum and functional analysis of PcHsp70-5 under drought stress
2024 Jul 8 ・A reference-grade genome of the xerophyte Ammopiptanthus mongolicus sheds light on its evolution history in legumes and drought-tolerance mechanisms
2024 Jan 23 ・Translocations and inversions: major chromosomal rearrangements during Vigna (Leguminosae) evolution
2023 Nov 21 ・Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response
2023 Dec 1 ・Unraveling the Unusual Subgenomic Organization in the Neopolyploid Free-Living Flatworm Macrostomum lignano
2023 Sep 3 ・Genes of Green Plants: Multiple Parallel Length Expansions, Intron Gains and Losses, Partial Gene Duplications, and Alternative Splicing
2023 Jul 31 ・Chromosome-level genome assembly and population genomics of Robinia pseudoacacia reveal the genetic basis for its wide cultivation
2023 Jun 20 ・Genome-wide identification and expression analysis of VQ gene family under abiotic stress in Coix lacryma-jobi L
2023 Apr 19 ・Two-step model of paleohexaploidy, ancestral genome reshuffling and plasticity of heat shock response in Asteraceae
2023 Jun 1 ・Genomic Insights into daptation to Karst Limestone and Incipient Speciation in East Asian Platycarya spp
2022 Dec ・Chromosome-length genome assemblies of six legume species provide insights into genome organization, evolution, and agronomic traits for crop improvement
2022 Sep 29 ・Genomic analyses of rice bean landraces reveal adaptation and yield related loci to accelerate breeding
2022 Nov 28 ・A common whole-genome paleotetraploidization in Cucurbitales. Plant Physiol
2022 Nov 14 ・Integrated network analyses identify MYB4R1 neofunctionalization in the UV-B adaptation of Tartary buckwheat
2022 May 31 ・Paleopolyploidies and Genomic Fractionation in Major Eudicot Clades
2022 Jun 17 ・Conversion between duplicated genes generated by polyploidization contributes to the divergence of poplar and willow
2022 Apr 20 ・PolyReco: A Method to Automatically Label Collinear Regions and Recognize Polyploidy Events Based on the Ks Dotplot
2022 May 27 ・Chromosome-level genome assembly and characterization of Sophora Japonica
2022 Apr 7 ・Reshuffling of the ancestral core-eudicot genome shaped chromatin topology and epigenetic modification in Panax
2022 Jul ・Genome sequence and population genomics provide insights into chromosomal evolution and phytochemical innovation of Hippophae rhamnoides
2021 Dec 1 ・Illegitimate Recombination between Duplicated Genes Generated from Recursive Polyploidizations Accelerated the Divergence of the Genus Arachis
2021 Dec 6 ・Genome-wide and expression analysis of B-box gene family in pepper
2021 Oct 26 ・Integrated Transcriptomic and Bioinformatics Analyses Reveal the Molecular Mechanisms for the Differences in Seed Oil and Starch Content Between Glycine max and Cicer arietinum
2021 Oct 26 ・Integrated Transcriptomic and Bioinformatics Analyses Reveal the Molecular Mechanisms for the Differences in Seed Oil and Starch Content Between Glycine max and Cicer arietinum
2021 Sep 7 ・A tetraploidization event shaped theAquilaria sinensis genome and contributed to the ability of sesquiterpenessynthesis
2022 Jan ・The genome sequenceprovides insights into salt tolerance of Achnatherum splendens (Gramineae), aconstructive species of alkaline grassland
2021 Jun 19 ・Conversionbetween 100-million-year-old duplicated genes contributes to rice subspeciesdivergence
2020 Oct 29 ・A multilayered cross-speciesanalysis of GRAS transcription factors uncovered their functional networks inplant adaptation to the environment
2021 Jan 6 ・Genome-wide identification and characterization of small auxin-up RNA (SAUR) genefamily in plants: evolution and expression profiles during normal growth andstress response
2020 Dec1 ・A chromosome-scale genome assembly of adiploid alfalfa, the progenitor of autotetraploid alfalfa
2020 Jun ・Paleo-polyploidization in Lycophytes
2021 Apr ・The celery genome sequence reveals sequential paleo-polyploidizations,karyotype evolution and resistance gene reduction in apiales
2020 Jul 21 ・Illegitimate Recombination Between Homeologous Genes in Wheat Genome
2020 Jul 6 ・Pervasive duplication, biased molecular evolution and comprehensive functionalanalysis of the PP2C family in Glycine max
2020 Jun 26 ・Transcriptome Analysis RevealsPotential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychisviciifolia to Extreme Environmental Conditions in the Qinghai-TibetanPlateau
2020 May 19 ・Allele-aware chromosome-level genome assemblyand efficient transgene-free genome editing for the autotetraploid cultivatedalfalfa
2020 Apr 23 ・CottonDuplicated Genes Produced by Polyploidy Show Significantly Elevated andUnbalanced Evolutionary Rates, Overwhelmingly Perturbing Gene Tree Topology
2020 Mar 8 ・Reconstruction of the EvolutionaryHistories of UGT Gene Superfamily in Legumes Clarifies the Functional Divergenceof Duplicates in Specialized Metabolism
2020 Feb 21 ・Preferentialgene retention increases the robustness of cold regulation in Brassicaceae and other plants after polyploidization
2020 Feb 4 ・Genome-wide identification andexpression analysis of the VQ gene family in Cicer arietinumand Medicago truncatula
2020 Sep ・Trait associations in the pangenome of pigeonpea (Cajanus cajan)
2020 Jan 31 ・Sequential Paleotetraploidization shaped the carrot genome
2020 Sep ・Genome-wide identification and expression analysis of the 14-3-3 genefamily in soybean (Glycine max)
2020 Jun ・Deciphering thehigh-quality genome sequence of coriander that causes controversial feelings
2019 Nov 14 ・The wax gourdgenomes offer insights into the genetic diversity and ancestral cucurbitkaryotype
2019 Sep 10 ・Polyploidy Index and Its Implications forthe Evolution of Polyploids
2019 Aug 21 ・Genome-wide identification and expression analysis of the VQ genefamily in soybean (Glycine max)
2019 Aug 6 ・Alignment of Rutaceae Genomes Reveals Lower Genome Fractionation Level ThanEudicot Genomes Affected by Extra Polyploidization
2019 Nov ・Development of gene-based molecular markerstagging low alkaloid pauper locus in white lupin (Lupinus albus L.)
2019 Feb 11 ・Paleogenomics:reconstruction of plant evolutionary trajectories from modern and ancient DNA
2019 Jan ・Recursive Paleohexaploidization Shaped the Durian Genome
2019 Feb 1 ・Landscape of gene transposition-duplication within theBrassicaceae family
2018 Oct 17 ・WRKY transcription factors in legumes
2018 Sep 28 ・Two Likely Auto-Tetraploidization Events ShapedKiwifruit Genome and Contributed to Establishment of the Actinidiaceae Family
2018 Sep 8 ・Physiology BasedApproaches for Breeding of Next-Generation Food Legumes
2018 Jun 14 ・Investigation of BaselineIron Levels in Australian Chickpea and Evaluation of a TransgenicBiofortification Approach
2017 Nov 30 ・Non-Additive Transcriptomic Responses to Inoculationwith Rhizobia in a Young Allopolyploid Compared with Its Diploid Progenitors.Genes (Basel)
2017 Dec ・Patterns and Consequences of SubgenomeDifferentiation Provide Insights into the Nature of Paleopolyploidy in Plants
2018 Jan 1 ・An OverlookedPaleotetraploidization in Cucurbitaceae
2017 Sep 2 ・Oil body biogenesis and biotechnology in legumeseeds